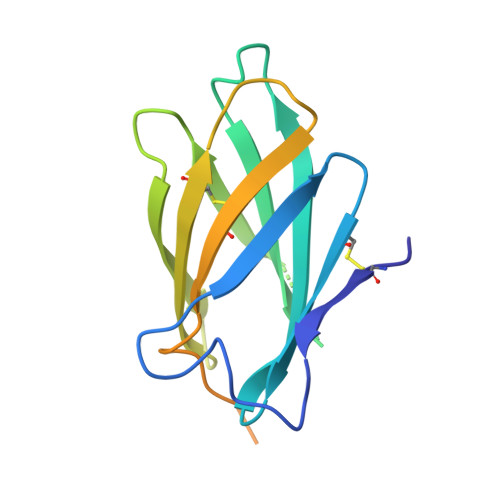
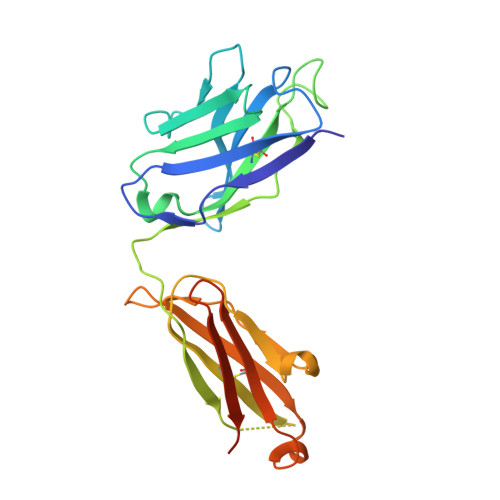
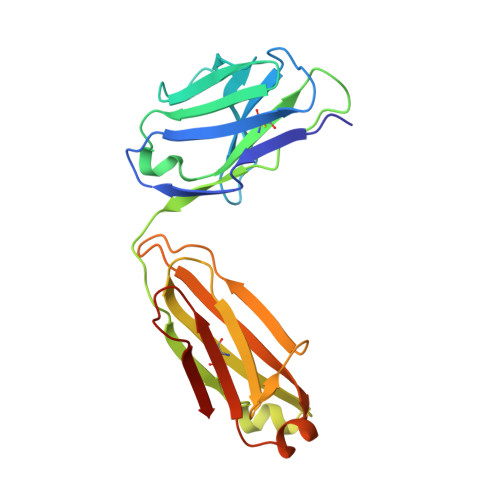
Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus.
Cockburn, J.J., Navarro Sanchez, M.E., Goncalvez, A.P., Zaitseva, E., Stura, E.A., Kikuti, C.M., Duquerroy, S., Dussart, P., Chernomordik, L.V., Lai, C.J., Rey, F.A.(2012) EMBO J 31: 767-779
- PubMed: 22139356
- DOI: https://doi.org/10.1038/emboj.2011.439
- Primary Citation of Related Structures:
3UAJ, 3UC0 - PubMed Abstract:
The four serotypes of dengue virus (DENV-1 to -4) cause the most important emerging viral disease. Protein E, the principal viral envelope glycoprotein, mediates fusion of the viral and endosomal membranes during virus entry and is the target of neutralizing antibodies. However, the epitopes of strongly neutralizing human antibodies have not been described despite their importance to vaccine development. The chimpanzee Mab 5H2 potently neutralizes DENV-4 by binding to domain I of E. The crystal structure of Fab 5H2 bound to E from DENV-4 shows that antibody binding prevents formation of the fusogenic hairpin conformation of E, which together with in-vitro assays, demonstrates that 5H2 neutralizes by blocking membrane fusion in the endosome. Furthermore, we show that human sera from patients recovering from DENV-4 infection contain antibodies that bind to the 5H2 epitope region on domain I. This study, thus, provides new information and tools for effective vaccine design to prevent dengue disease.
- Département de Virologie, Institut Pasteur, Unité de Virologie Structurale, Paris, France. joseph.cockburn@cancer.org.uk
Organizational Affiliation: