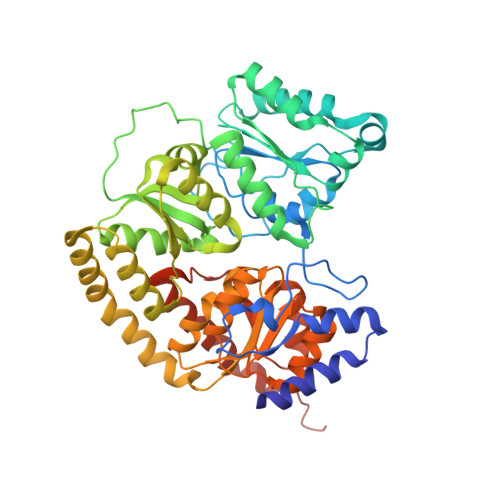
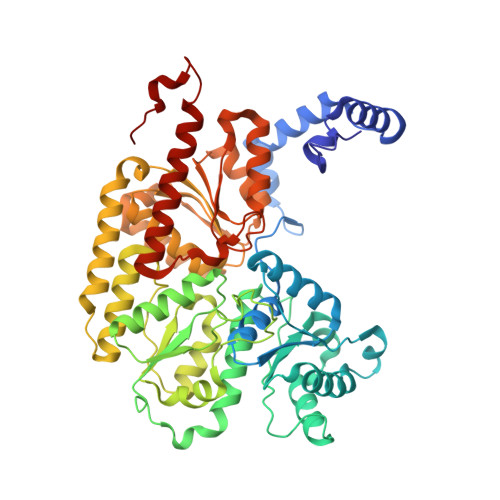
Evidence for interstitial carbon in nitrogenase FeMo cofactor.
Spatzal, T., Aksoyoglu, M., Zhang, L., Andrade, S.L., Schleicher, E., Weber, S., Rees, D.C., Einsle, O.(2011) Science 334: 940-940
- PubMed: 22096190
- DOI: https://doi.org/10.1126/science.1214025
- Primary Citation of Related Structures:
3U7Q - PubMed Abstract:
The identity of the interstitial light atom in the center of the FeMo cofactor of nitrogenase has been enigmatic since its discovery. Atomic-resolution x-ray diffraction data and an electron spin echo envelope modulation (ESEEM) analysis now provide direct evidence that the ligand is a carbon species.
- Institut für Organische Chemie und Biochemie, Albert-Ludwigs-Universität Freiburg, Freiburg, Germany.
Organizational Affiliation: