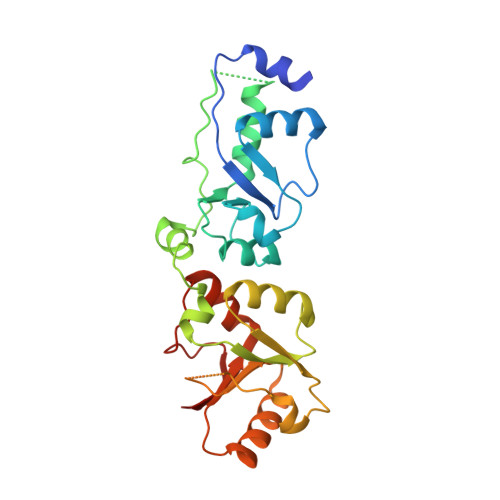
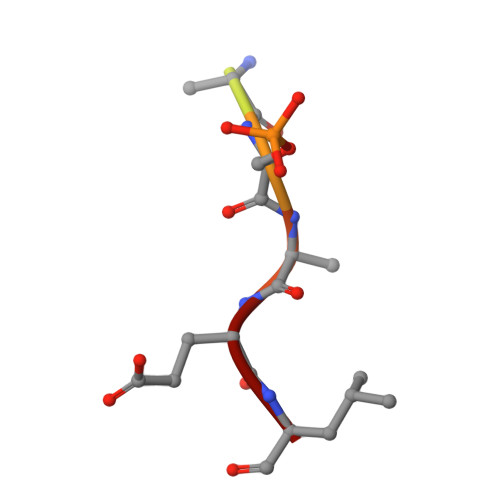
Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
Li, X., Liu, K., Li, F., Wang, J., Huang, H., Wu, J., Shi, Y.(2012) J Biological Chem 287: 9137-9146
- PubMed: 22262834
- DOI: https://doi.org/10.1074/jbc.M111.311860
- Primary Citation of Related Structures:
3T7I, 3T7J, 3T7K - PubMed Abstract:
Rtt107 (regulator of Ty1 transposition 107; Esc4) is a DNA repair protein from Saccharomyces cerevisiae that can restore stalled replication forks following DNA damage. There are six BRCT (BRCA1 C-terminal) domains in Rtt107 that act as binding sites for other recruited proteins during DNA repair. Several Rtt107 binding partners have been identified, including Slx4, Rtt101, Rad55, and the Smc5/6 (structural maintenance of chromosome) protein complex. Rtt107 can reportedly be recruited to chromatin in the presence of Rtt101 and Rtt109 upon DNA damage, but the chromatin-binding site of Rtt107 has not been identified. Here, we report our investigation of the interaction between phosphorylated histone H2A (γH2A) and the C-terminal tandem BRCT repeats (BRCT(5)-BRCT(6)) of Rtt107. The crystal structures of BRCT(5)-BRCT(6) alone and in a complex with γH2A reveal the molecular basis of the Rtt107-γH2A interaction. We used in vitro mutagenesis and a fluorescence polarization assay to confirm the location of the Rtt107 motif that is crucial for this interaction. In addition, these assays indicated that this interaction requires the phosphorylation of H2A. An in vivo phenotypic analysis in yeast demonstrated the critical role of BRCT(5)-BRCT(6) and its interaction with γH2A during the DNA damage response. Our results shed new light on the molecular mechanism by which Rtt107 is recruited to chromatin in response to stalled DNA replication forks.
- Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, China.
Organizational Affiliation: