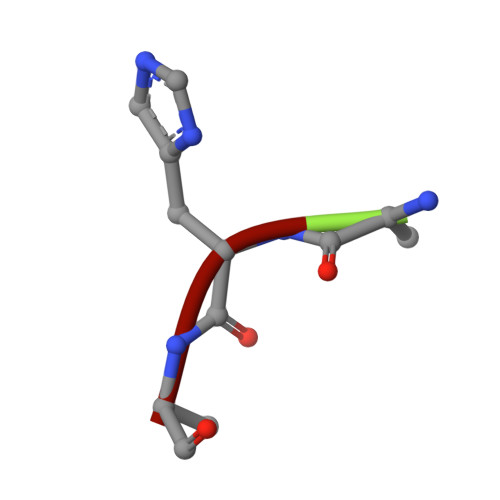
Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Hampton, S.E., Baragana, B., Schipani, A., Bosch-Navarrete, C., Musso-Buendia, J.A., Recio, E., Kaiser, M., Whittingham, J.L., Roberts, S.M., Shevtsov, M., Brannigan, J.A., Kahnberg, P., Brun, R., Wilson, K.S., Gonzalez-Pacanowska, D., Johansson, N.G., Gilbert, I.H.(2011) ChemMedChem 6: 1816-1831
- PubMed: 22049550
- DOI: https://doi.org/10.1002/cmdc.201100255
- Primary Citation of Related Structures:
3T60, 3T64, 3T6Y, 3T70 - PubMed Abstract:
Deoxyuridine 5'-triphosphate nucleotidohydrolase (dUTPase) is a potential drug target for malaria. We previously reported some 5'-tritylated deoxyuridine analogues (both cyclic and acyclic) as selective inhibitors of the Plasmodium falciparum dUTPase. Modelling studies indicated that it might be possible to replace the trityl group with a diphenyl moiety, as two of the phenyl groups are buried, whereas the third is exposed to solvent. Herein we report the synthesis and evaluation of some diphenyl analogues that have lower lipophilicity and molecular weight than the trityl lead compound. Co-crystal structures show that the diphenyl inhibitors bind in a similar manner to the corresponding trityl derivatives, with the two phenyl moieties occupying the predicted buried phenyl binding sites. The diphenyl compounds prepared show similar or slightly lower inhibition of PfdUTPase, and similar or weaker inhibition of parasite growth than the trityl compounds.
- Division of Biological Chemistry and Drug Discovery, College of Life Science, University of Dundee, Sir James Black Centre, Dundee DD1 5EH, UK.
Organizational Affiliation: