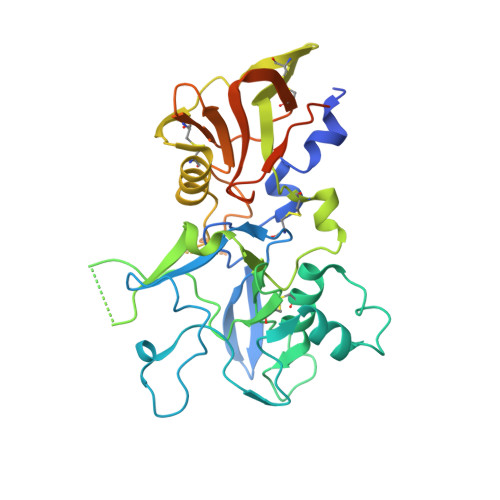
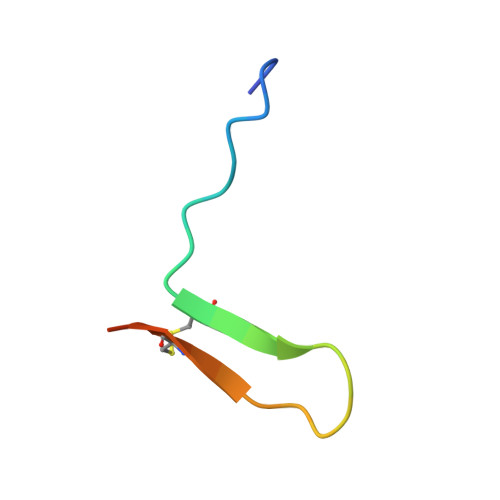
Structural and functional insights into the malaria parasite moving junction complex.
Vulliez-Le Normand, B., Tonkin, M.L., Lamarque, M.H., Langer, S., Hoos, S., Roques, M., Saul, F.A., Faber, B.W., Bentley, G.A., Boulanger, M.J., Lebrun, M.(2012) PLoS Pathog 8: e1002755-e1002755
- PubMed: 22737069
- DOI: https://doi.org/10.1371/journal.ppat.1002755
- Primary Citation of Related Structures:
3SRI, 3SRJ, 3ZWZ - PubMed Abstract:
Members of the phylum Apicomplexa, which include the malaria parasite Plasmodium, share many features in their invasion mechanism in spite of their diverse host cell specificities and life cycle characteristics. The formation of a moving junction (MJ) between the membranes of the invading apicomplexan parasite and the host cell is common to these intracellular pathogens. The MJ contains two key parasite components: the surface protein Apical Membrane Antigen 1 (AMA1) and its receptor, the Rhoptry Neck Protein (RON) complex, which is targeted to the host cell membrane during invasion. In particular, RON2, a transmembrane component of the RON complex, interacts directly with AMA1. Here, we report the crystal structure of AMA1 from Plasmodium falciparum in complex with a peptide derived from the extracellular region of PfRON2, highlighting clear specificities of the P. falciparum RON2-AMA1 interaction. The receptor-binding site of PfAMA1 comprises the hydrophobic groove and a region that becomes exposed by displacement of the flexible Domain II loop. Mutations of key contact residues of PfRON2 and PfAMA1 abrogate binding between the recombinant proteins. Although PfRON2 contacts some polymorphic residues, binding studies with PfAMA1 from different strains show that these have little effect on affinity. Moreover, we demonstrate that the PfRON2 peptide inhibits erythrocyte invasion by P. falciparum merozoites and that this strong inhibitory potency is not affected by AMA1 polymorphisms. In parallel, we have determined the crystal structure of PfAMA1 in complex with the invasion-inhibitory peptide R1 derived by phage display, revealing an unexpected structural mimicry of the PfRON2 peptide. These results identify the key residues governing the interactions between AMA1 and RON2 in P. falciparum and suggest novel approaches to antimalarial therapeutics.
Organizational Affiliation:
Unité d'Immunologie Structurale, Institut Pasteur, Paris, France.