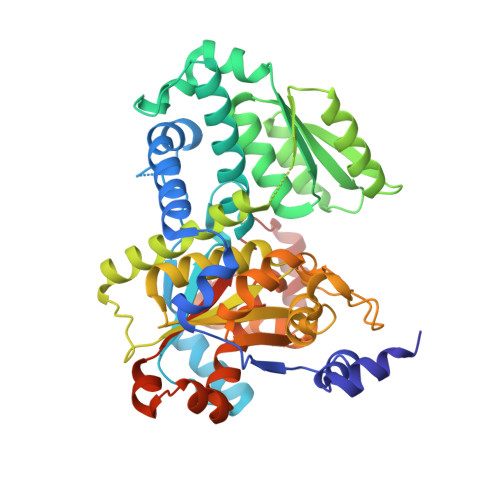
Crystal structures of open and closed forms of D-serine deaminase from Salmonella typhimurium - implications on substrate specificity and catalysis
Bharath, S.R., Bisht, S., Savithri, H.S., Murthy, M.R.N.(2011) FEBS J
- PubMed: 21668644
- DOI: https://doi.org/10.1111/j.1742-4658.2011.08210.x
- Primary Citation of Related Structures:
3R0X, 3R0Z - PubMed Abstract:
Metabolism of D-amino acids is of considerable interest due to their key importance in cell structure and function. Salmonella typhimuriumd-serine deaminase (StDSD) is a pyridoxal 5' phosphate (PLP) dependent enzyme that catalyses degradation of D-Ser to pyruvate and ammonia. The first crystal structure of d-serine deaminase described here reveals a typical Foldtype II or tryptophan synthase β subunit fold of PLP-dependent enzymes. Although holoenzyme was used for crystallization of both wild-type StDSD (WtDSD) and selenomethionine labelled StDSD (SeMetDSD), significant electron density was not observed for the cofactor, indicating that the enzyme has a low affinity for the cofactor under crystallization conditions. Interestingly, unexpected conformational differences were observed between the two structures. The WtDSD was in an open conformation while SeMetDSD, crystallized in the presence of isoserine, was in a closed conformation suggesting that the enzyme is likely to undergo conformational changes upon binding of substrate as observed in other Foldtype II PLP-dependent enzymes. Electron density corresponding to a plausible sodium ion was found near the active site of the closed but not in the open state of the enzyme. Examination of the active site and substrate modelling suggests that Thr166 may be involved in abstraction of proton from the Cα atom of the substrate. Apart from the physiological reaction, StDSD catalyses α, β elimination of D-Thr, D-Allothr and L-Ser to the corresponding α-keto acids and ammonia. The structure of StDSD provides a molecular framework necessary for understanding differences in the rate of reaction with these substrates.
- Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India.
Organizational Affiliation: