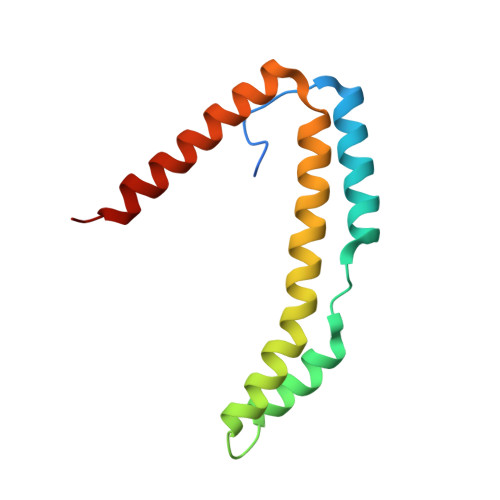
Structure of the Periplasmic Stress Response Protein CpxP.
Thede, G.L., Arthur, D.C., Edwards, R.A., Buelow, D.R., Wong, J.L., Raivio, T.L., Glover, J.N.(2011) J Bacteriol 193: 2149-2157
- PubMed: 21317318
- DOI: https://doi.org/10.1128/JB.01296-10
- Primary Citation of Related Structures:
3QZC - PubMed Abstract:
CpxP is a novel bacterial periplasmic protein with no homologues of known function. In gram-negative enteric bacteria, CpxP is thought to interact with the two-component sensor kinase, CpxA, to inhibit induction of the Cpx envelope stress response in the absence of protein misfolding. CpxP has also been shown to facilitate DegP-mediated proteolysis of misfolded proteins. Six mutations that negate the ability of CpxP to function as a signaling protein are localized in or near two conserved LTXXQ motifs that define a class of proteins with similarity to CpxP, Pfam PF07813. To gain insight into how these mutations might affect CpxP signaling and/or proteolytic adaptor functions, the crystal structure of CpxP from Escherichia coli was determined to 2.85-Å resolution. The structure revealed an antiparallel dimer of intertwined α-helices with a highly basic concave surface. Each protomer consists of a long, hooked and bent hairpin fold, with the conserved LTXXQ motifs forming two diverging turns at one end. Biochemical studies demonstrated that CpxP maintains a dimeric state but may undergo a slight structural adjustment in response to the inducing cue, alkaline pH. Three of the six previously characterized cpxP loss-of-function mutations, M59T, Q55P, and Q128H, likely result from a destabilization of the protein fold, whereas the R60Q, D61E, and D61V mutations may alter intermolecular interactions.
Organizational Affiliation:
Department of Biochemistry, School of Molecular and Systems Medicine, University of Alberta, Edmonton, Alberta, Canada.