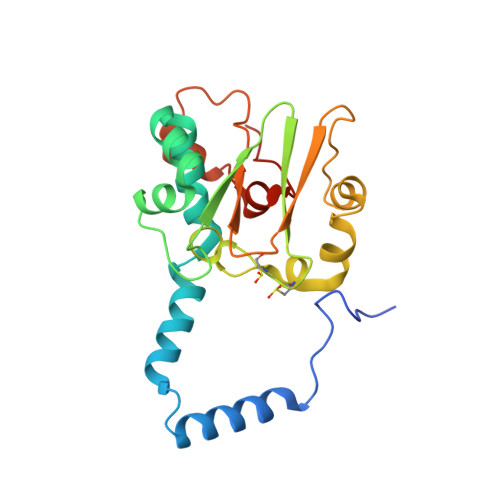
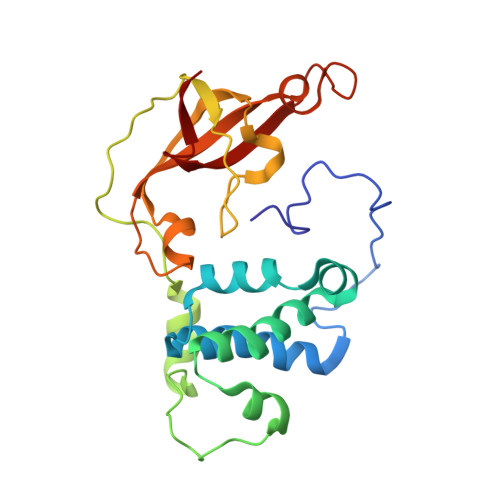
Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Brodkin, H.R., Novak, W.R., Milne, A.C., D'Aquino, J.A., Karabacak, N.M., Goldberg, I.G., Agar, J.N., Payne, M.S., Petsko, G.A., Ondrechen, M.J., Ringe, D.(2011) Biochemistry 50: 4923-4935
- PubMed: 21473592
- DOI: https://doi.org/10.1021/bi101761e
- Primary Citation of Related Structures:
3QXE, 3QYG, 3QYH, 3QZ5, 3QZ9 - PubMed Abstract:
Active sites may be regarded as layers of residues, whereby the residues that interact directly with substrate also interact with residues in a second shell and these in turn interact with residues in a third shell. These residues in the second and third layers may have distinct roles in maintaining the essential chemical properties of the first-shell catalytic residues, particularly their spatial arrangement relative to the substrate binding pocket, and their electrostatic and dynamic properties. The extent to which these remote residues participate in catalysis and precisely how they affect first-shell residues remains unexplored. To improve our understanding of the roles of second- and third-shell residues in catalysis, we used THEMATICS to identify residues in the second and third shells of the Co-type nitrile hydratase from Pseudomonas putida (ppNHase) that may be important for catalysis. Five of these predicted residues, and three additional, conserved residues that were not predicted, have been conservatively mutated, and their effects have been studied both kinetically and structurally. The eight residues have no direct contact with the active site metal ion or bound substrate. These results demonstrate that three of the predicted second-shell residues (α-Asp164, β-Glu56, and β-His147) and one predicted third-shell residue (β-His71) have significant effects on the catalytic efficiency of the enzyme. One of the predicted residues (α-Glu168) and the three residues not predicted (α-Arg170, α-Tyr171, and β-Tyr215) do not have any significant effects on the catalytic efficiency of the enzyme.
- Department of Chemistry and Chemical Biology and Institute for Complex Scientific Software, Northeastern University, Boston, Massachusetts 02115, USA.
Organizational Affiliation: