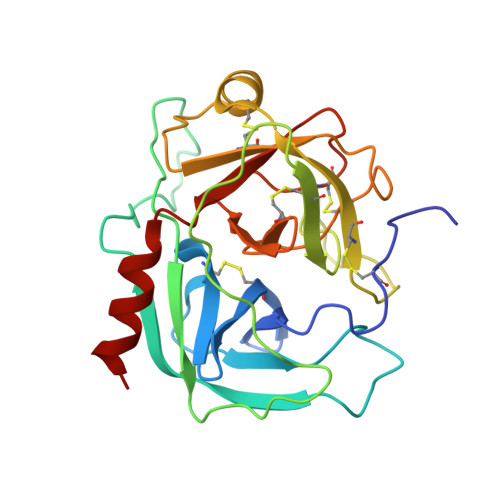
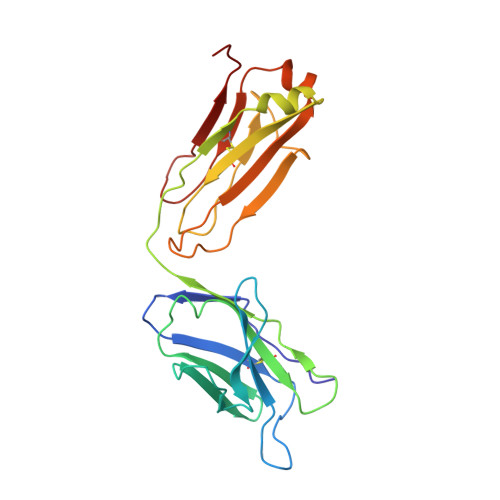
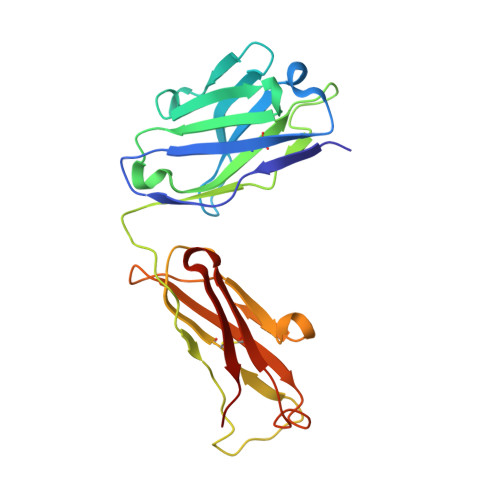
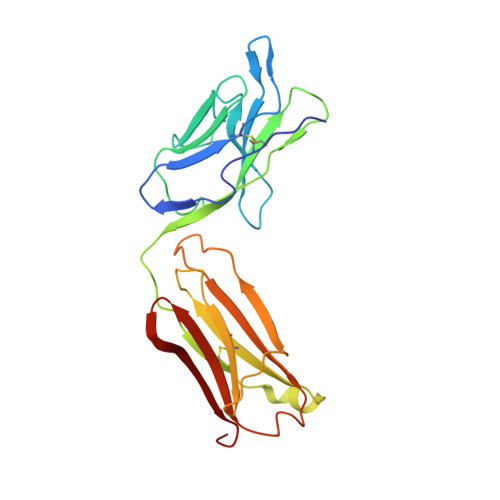
Crystal structure of human prostate-specific antigen in a sandwich antibody complex.
Stura, E.A., Muller, B.H., Bossus, M., Michel, S., Jolivet-Reynaud, C., Ducancel, F.(2011) J Mol Biology 414: 530-544
- PubMed: 22037582
- DOI: https://doi.org/10.1016/j.jmb.2011.10.007
- Primary Citation of Related Structures:
3QUM - PubMed Abstract:
Human prostate-specific antigen (PSA or human kallikrein-related peptidase 3) present in small quantities in the sera of healthy men becomes elevated in prostate cancer (PCa) and other prostate disorders. The ability to identify the free PSA fraction associated with PCa could increase the reliability of the PSA diagnostic test. Here we present the crystal structure of human PSA from seminal fluid in a sandwich complex with two monoclonal antibodies (mAbs). MAb 5D5A5 captures total PSA with exceptionally high affinity, and mAb 5D3D11 selectively discriminates between free PSA subforms that are more abundant in sera from patients with PCa. Although the antigen is not of seric origin, several insights into cancer diagnosis can be discerned from this complex. MAb 5D3D11 recognizes a PSA conformation different from that previously reported. Interacting with the kallikrein loop, the PSA N-linked glycan attached to asparagine 61 is an uncommonly complex sialated triantennary chain. O-linked glycosylation is observed at threonine 125. The description of how PSA subforms in prostatic fluid can be discriminated using pairs of antibodies is a first step in the design of new strategies that are capable of real discrimination among PSA subforms, which will lead to the formulation of more reliable diagnostic tests. In a companion article [Muller, B. H., Savatier, A., L'Hostis, G., Costa, N., Bossus, M., Michel, S., et al. (2011). In vitro affinity maturation of an anti-PSA antibody for prostate cancer diagnostic assay. J. Mol. Biol.], we describe engineering efforts to improve the affinity of mAb 5D3D11, a first step towards such goal.
- CEA, iBiTec-S, Service d'Ingénierie Moléculaire des Protéines, Laboratoire de Toxinologie Moléculaire et Biotechnologies, Gif-sur-Yvette F-91191, France. estura@cea.fr
Organizational Affiliation: