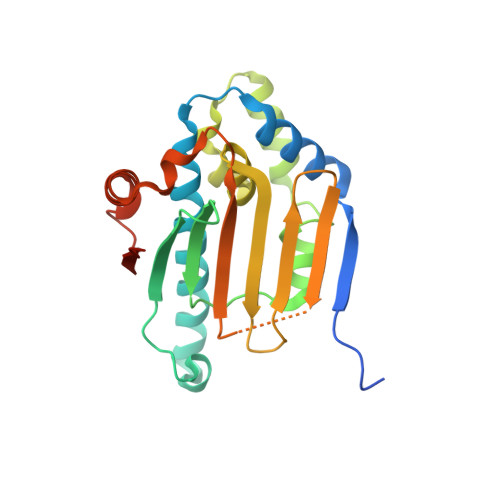
Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin.
Wernimont, A.K., Tempel, W., Lin, Y.H., Hutchinson, A., MacKenzie, F., Fairlamb, A., Cossar, D., Zhao, Y., Schapira, M., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Weigelt, J., Ferguson, M.A.J., Hui, R., Pizarro, J.C., Hills, T.To be published.