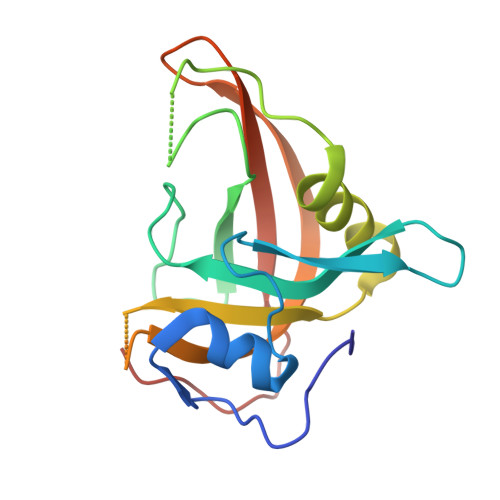
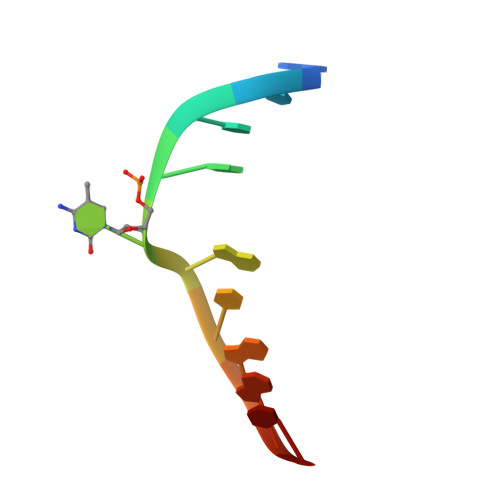
A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Rajakumara, E., Law, J.A., Simanshu, D.K., Voigt, P., Johnson, L.M., Reinberg, D., Patel, D.J., Jacobsen, S.E.(2011) Genes Dev 25: 137-152
- PubMed: 21245167
- DOI: https://doi.org/10.1101/gad.1980311
- Primary Citation of Related Structures:
3Q0B, 3Q0C, 3Q0D, 3Q0F - PubMed Abstract:
Cytosine DNA methylation is evolutionarily ancient, and in eukaryotes this epigenetic modification is associated with gene silencing. Proteins with SRA (SET- or RING-associated) methyl-binding domains are required for the establishment and/or maintenance of DNA methylation in both plants and mammals. The 5-methyl-cytosine (5mC)-binding specificity of several SRA domains have been characterized, and each one has a preference for DNA methylation in different sequence contexts. Here we demonstrate through mobility shift assays and calorimetric measurements that the SU(VAR)3-9 HOMOLOG 5 (SUVH5) SRA domain differs from other SRA domains in that it can bind methylated DNA in all contexts to similar extents. Crystal structures of the SUVH5 SRA domain bound to 5mC-containing DNA in either the fully or hemimethylated CG context or the methylated CHH context revealed a dual flip-out mechanism where both the 5mC and a base (5mC, C, or G, respectively) from the partner strand are simultaneously extruded from the DNA duplex and positioned within binding pockets of individual SRA domains. Our structure-based in vivo studies suggest that a functional SUVH5 SRA domain is required for both DNA methylation and accumulation of the H3K9 dimethyl modification in vivo, suggesting a role for the SRA domain in recruitment of SUVH5 to genomic loci.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.
Organizational Affiliation: