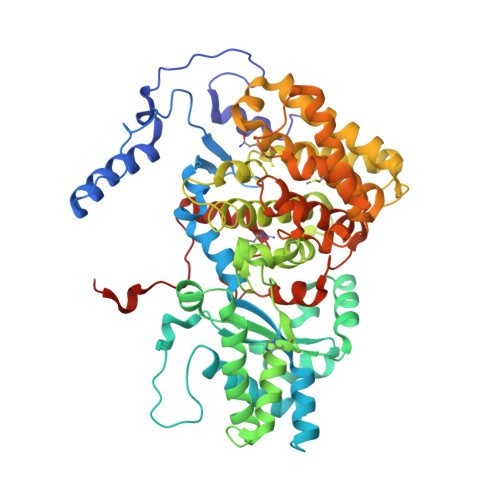
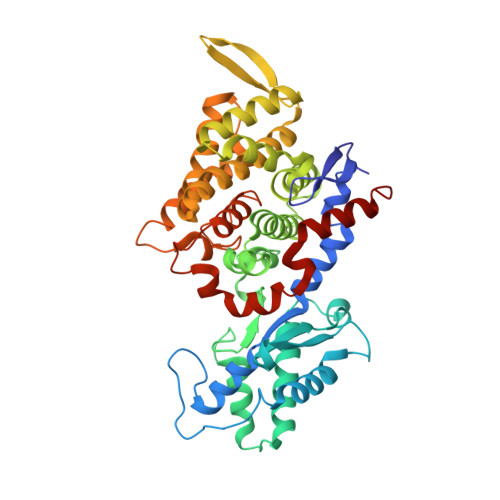
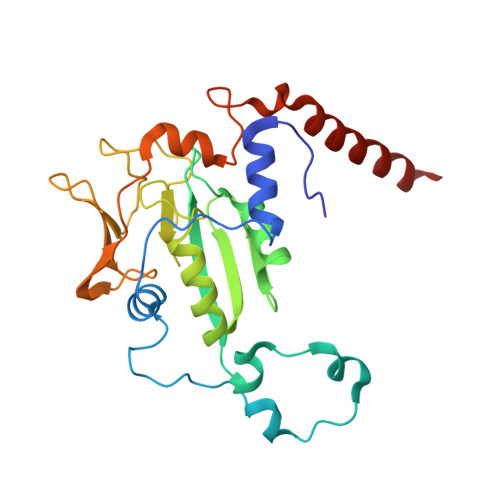
Structural Analysis of a Ni-Methyl Species in Methyl-Coenzyme M Reductase from Methanothermobacter marburgensis.
Cedervall, P.E., Dey, M., Li, X., Sarangi, R., Hedman, B., Ragsdale, S.W., Wilmot, C.M.(2011) J Am Chem Soc 133: 5626-5628
- PubMed: 21438550
- DOI: https://doi.org/10.1021/ja110492p
- Primary Citation of Related Structures:
3POT - PubMed Abstract:
We present the 1.2 Å resolution X-ray crystal structure of a Ni-methyl species that is a proposed catalytic intermediate in methyl-coenzyme M reductase (MCR), the enzyme that catalyzes the biological formation of methane. The methyl group is situated 2.1 Å proximal of the Ni atom of the MCR coenzyme F(430). A rearrangement of the substrate channel has been posited to bring together substrate species, but Ni(III)-methyl formation alone does not lead to any observable structural changes in the channel.
- Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Organizational Affiliation: