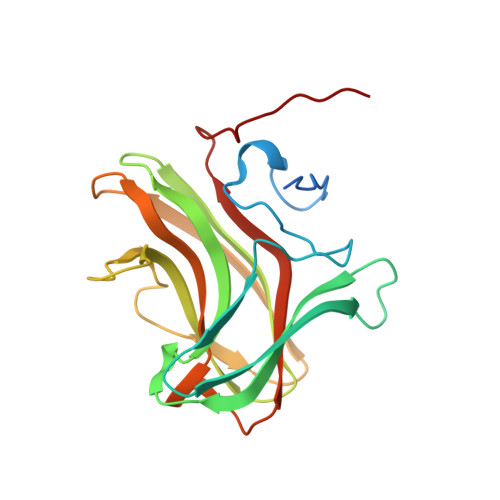
Structure of CBM4 from Clostridium thermocellum cellulase K.
Alahuhta, M., Luo, Y., Ding, S.Y., Himmel, M.E., Lunin, V.V.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 527-530
- PubMed: 21543854
- DOI: https://doi.org/10.1107/S1744309111003307
- Primary Citation of Related Structures:
3P6B - PubMed Abstract:
Here, a 2.0 Å resolution X-ray structure of Clostridium thermocellum cellulase K family 4 carbohydrate-binding module (CelK CBM4) is reported. The resulting structure was refined to an R factor of 0.212 and an R(free) of 0.274. Structural analysis shows that this new structure is very similar to the previously solved structure of C. thermocellum CbhA CBM4. Most importantly, these data support the previously proposed notion of an extended binding pocket using a novel tryptophan-containing loop that may be highly conserved in clostridial CBM4 proteins.
- BioSciences Center, National Renewable Energy Laboratory, 1617 Cole Boulevard, Golden, Colorado 80401-3305, USA.
Organizational Affiliation: