Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr kinase, PknB
Lombana, T.N., Echols, N., Good, M.C., Thomsen, N.D., Ng, H.-L., Greenstein, A.E., Falick, A.M., King, D.S., Alber, T.(2010) Structure 18: 1667-1677
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2010) Structure 18: 1667-1677
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine/threonine protein kinase | 311 | Mycobacterium tuberculosis H37Ra | Mutation(s): 1 Gene Names: MRA_0016, pknB, PknB (Rv0014c) EC: 2.7.11.1 | 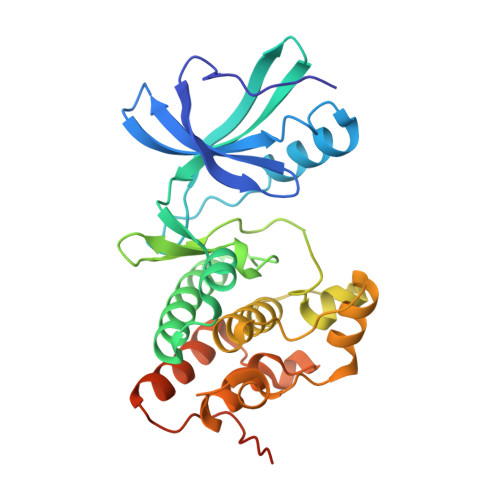 | |
UniProt | |||||
Find proteins for P9WI81 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WI81 Go to UniProtKB: P9WI81 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WI81 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AGS Query on AGS | B [auth A] | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER C10 H16 N5 O12 P3 S NLTUCYMLOPLUHL-KQYNXXCUSA-N |  | ||
| TRS Query on TRS | E [auth A] | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C4 H12 N O3 LENZDBCJOHFCAS-UHFFFAOYSA-O |  | ||
| SO4 Query on SO4 | D [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| MN Query on MN | C [auth A] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 39.341 | α = 90 |
| b = 51.603 | β = 90 |
| c = 155.126 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Blu-Ice | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |