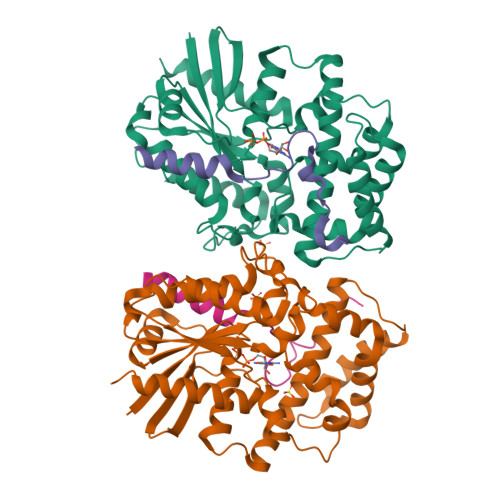
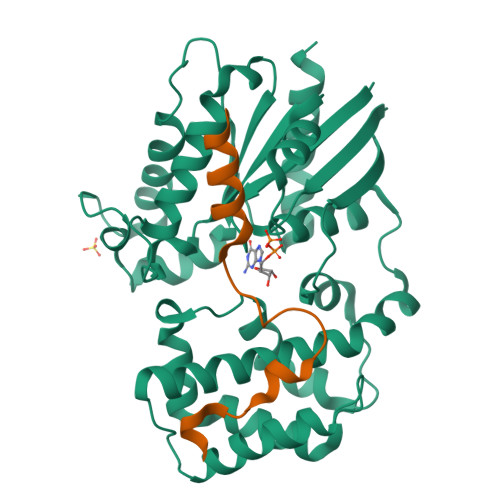
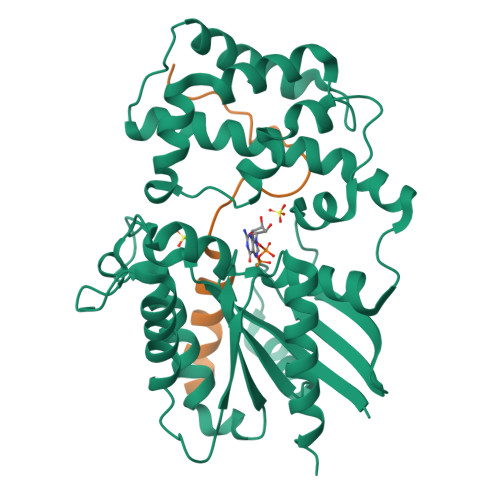
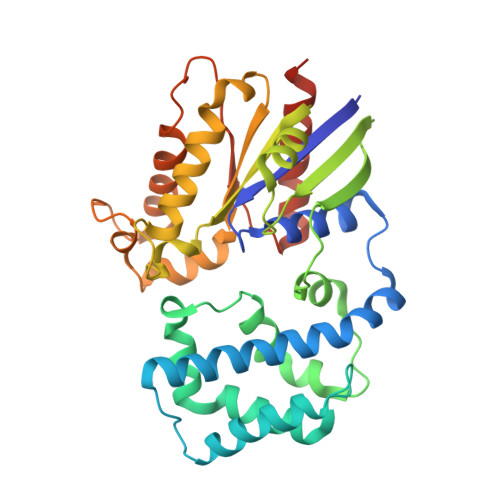
Structural Determinants of Affinity Enhancement between GoLoco Motifs and G-Protein {alpha} Subunit Mutants.
Bosch, D.E., Kimple, A.J., Sammond, D.W., Muller, R.E., Miley, M.J., Machius, M., Kuhlman, B., Willard, F.S., Siderovski, D.P.(2011) J Biological Chem 286: 3351-3358
- PubMed: 21115486
- DOI: https://doi.org/10.1074/jbc.M110.190496
- Primary Citation of Related Structures:
3ONW - PubMed Abstract:
GoLoco motif proteins bind to the inhibitory G(i) subclass of G-protein α subunits and slow the release of bound GDP; this interaction is considered critical to asymmetric cell division and neuro-epithelium and epithelial progenitor differentiation. To provide protein tools for interrogating the precise cellular role(s) of GoLoco motif/Gα(i) complexes, we have employed structure-based protein design strategies to predict gain-of-function mutations that increase GoLoco motif binding affinity. Here, we describe fluorescence polarization and isothermal titration calorimetry measurements showing three predicted Gα(i1) point mutations, E116L, Q147L, and E245L; each increases affinity for multiple GoLoco motifs. A component of this affinity enhancement results from a decreased rate of dissociation between the Gα mutants and GoLoco motifs. For Gα(i1)(Q147L), affinity enhancement was seen to be driven by favorable changes in binding enthalpy, despite reduced contributions from binding entropy. The crystal structure of Gα(i1)(Q147L) bound to the RGS14 GoLoco motif revealed disorder among three peptide residues surrounding a well defined Leu-147 side chain. Monte Carlo simulations of the peptide in this region showed a sampling of multiple backbone conformations in contrast to the wild-type complex. We conclude that mutation of Glu-147 to leucine creates a hydrophobic surface favorably buried upon GoLoco peptide binding, yet the hydrophobic Leu-147 also promotes flexibility among residues 511-513 of the RGS14 GoLoco peptide.
Organizational Affiliation:
Department of Pharmacology, University of North Carolina, Chapel Hill, North Carolina 27599, USA.