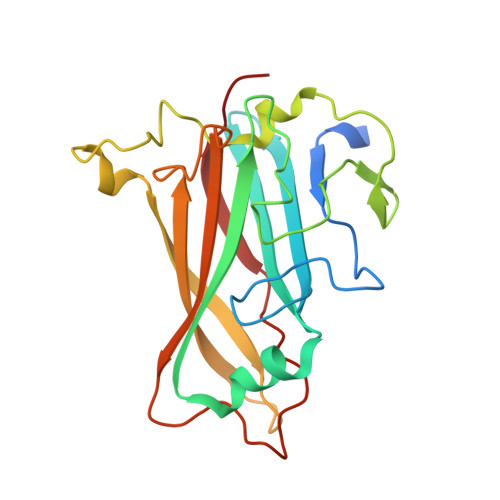
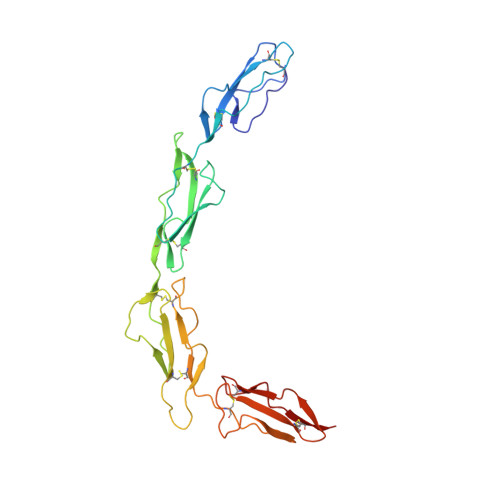
Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens.
Persson, B.D., Schmitz, N.B., Santiago, C., Zocher, G., Larvie, M., Scheu, U., Casasnovas, J.M., Stehle, T.(2010) PLoS Pathog 6: e1001122-e1001122
- PubMed: 20941397
- DOI: https://doi.org/10.1371/journal.ppat.1001122
- Primary Citation of Related Structures:
3O8E - PubMed Abstract:
The human membrane cofactor protein (MCP, CD46) is a central component of the innate immune system. CD46 protects autologous cells from complement attack by binding to complement proteins C3b and C4b and serving as a cofactor for their cleavage. Recent data show that CD46 also plays a role in mediating acquired immune responses, and in triggering autophagy. In addition to these physiologic functions, a significant number of pathogens, including select adenoviruses, measles virus, human herpes virus 6 (HHV-6), Streptococci, and Neisseria, use CD46 as a cell attachment receptor. We have determined the crystal structure of the extracellular region of CD46 in complex with the human adenovirus type 11 fiber knob. Extracellular CD46 comprises four short consensus repeats (SCR1-SCR4) that form an elongated structure resembling a hockey stick, with a long shaft and a short blade. Domains SCR1, SCR2 and SCR3 are arranged in a nearly linear fashion. Unexpectedly, however, the structure reveals a profound bend between domains SCR3 and SCR4, which has implications for the interactions with ligands as well as the orientation of the protein at the cell surface. This bend can be attributed to an insertion of five hydrophobic residues in a SCR3 surface loop. Residues in this loop have been implicated in interactions with complement, indicating that the bend participates in binding to C3b and C4b. The structure provides an accurate framework for mapping all known ligand binding sites onto the surface of CD46, thereby advancing an understanding of how CD46 acts as a receptor for pathogens and physiologic ligands of the immune system.
- University of Tuebingen, Tuebingen, Germany.
Organizational Affiliation: