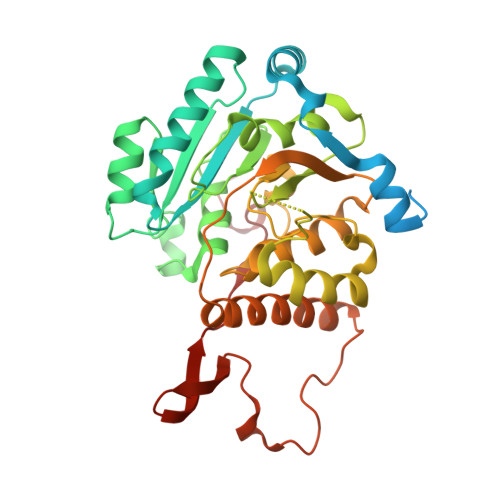
Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity
Empadinhas, N., Pereira, P.J.B., Albuquerque, L., Costa, J., Sa-Moura, B., Marques, A.T., Macedo-Ribeiro, S., da Costa, M.S.(2011) Mol Microbiol 79: 76-93
- PubMed: 21166895
- DOI: https://doi.org/10.1111/j.1365-2958.2010.07432.x
- Primary Citation of Related Structures:
3F1Y, 3KIA, 3O3P - PubMed Abstract:
Rubrobacter xylanophilus is the only actinobacterium known to accumulate the organic solute mannosylglycerate (MG); moreover, the accumulation of MG is constitutive. The key enzyme for MG synthesis, catalysing the conversion of GDP-mannose (GDP-Man) and D-3-phosphoglycerate (3-PGA) into the phosphorylated intermediate mannosyl-3-phosphoglycerate and GDP, was purified from R. xylanophilus cell extracts and the corresponding gene was expressed in E. coli. Despite the related solute glucosylglycerate (GG) having never been detected in R. xylanophilus, the cell extracts and the pure recombinant mannosyl-3-phosphoglycerate synthase (MpgS) could also synthesize glucosyl-3-phosphoglycerate (GPG), the precursor of GG, in agreement with the higher homology of the novel MpgS towards GPG-synthesizing mycobacterial glucosyl-3-phosphoglycerate synthases (GpgS) than towards MpgSs from hyper/thermophiles, known to accumulate MG under salt or thermal stress. To understand the specificity and substrate ambiguity of this novel enzyme, we determined the crystal structure of the unliganded MpgS and of its complexes with the nucleotide and sugar donors, at 2.2, 2.8 and 2.5 Å resolution respectively. The first three-dimensional structures of a protein from this extremely gamma-radiation-resistant thermophile here reported show that MpgS (GT81 family) contains a GT-A like fold and clearly explain its nucleotide and sugar-donor specificity. In the GDP-Man complex, a flexible loop ((254) RQNRHQ(259) ), located close to the active site moves towards the incoming sugar moiety, providing the ligands for both magnesium ion co-ordination and sugar binding. A triple mutant of R. xylanophilus MpgS, mimicking the (206) PLAGE(210) loop stabilizing hydrogen bond network observed for mycobacterial GpgSs, reduces significantly the affinity to GDP-Man, implicating this loop in the sugar-donor discrimination.
- Center for Neuroscience and Cell Biology, University of Coimbra, 3004-517 Coimbra, Portugal.
Organizational Affiliation: