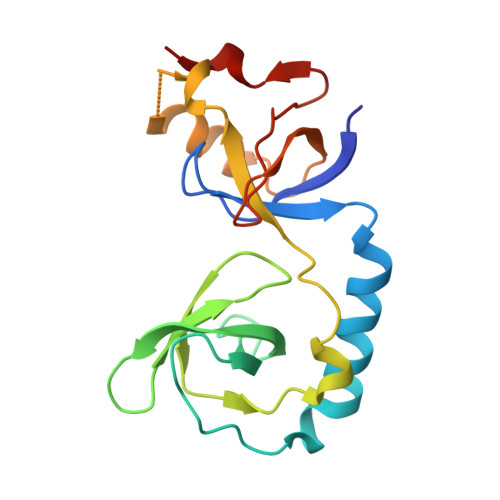
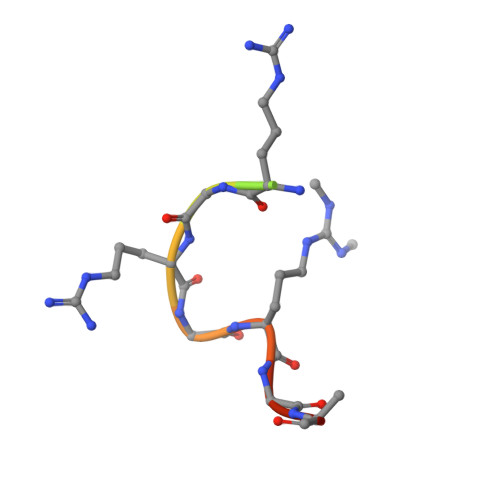
Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Liu, H.P., Wang, J.Y., Huang, Y., Li, Z.Z., Gong, W.M., Lehmann, R., Xu, R.M.(2010) Genes Dev 24: 1876-1881
- PubMed: 20713507
- DOI: https://doi.org/10.1101/gad.1956010
- Primary Citation of Related Structures:
3NTH, 3NTI, 3NTK - PubMed Abstract:
Piwi proteins are modified by symmetric dimethylation of arginine (sDMA), and the methylarginine-dependent interaction with Tudor domain proteins is critical for their functions in germline development. Cocrystal structures of an extended Tudor domain (eTud) of Drosophila Tudor with methylated peptides of Aubergine, a Piwi family protein, reveal that sDMA is recognized by an asparagine-gated aromatic cage. Furthermore, the unexpected Tudor-SN/p100 fold of eTud is important for sensing the position of sDMA. The structural information provides mechanistic insights into sDMA-dependent Piwi-Tudor interaction, and the recognition of sDMA by Tudor domains in general.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.
Organizational Affiliation: