Mechanism of molecular specificity of RhoGAP domains towards small GTPases of RhoA family.
Utepbergenov, D., Cooper, D.R., Derewenda, U., Somlyo, A.V., Derewenda, Z.S.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transforming protein RhoA | 180 | Homo sapiens | Mutation(s): 1 Gene Names: ARH12, ARHA, RHO12, RHOA EC: 3.6.5.2 | 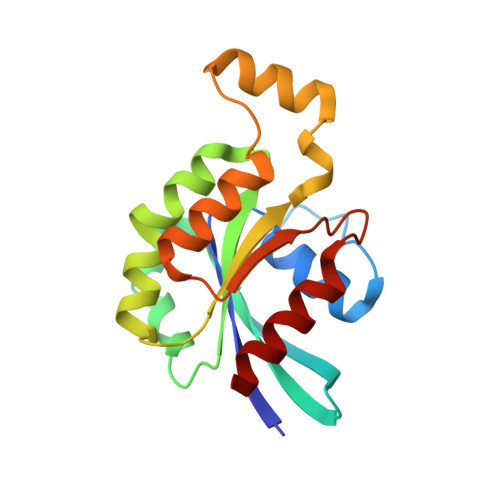 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61586 (Homo sapiens) Explore P61586 Go to UniProtKB: P61586 | |||||
PHAROS: P61586 GTEx: ENSG00000067560 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61586 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Rho GTPase-activating protein 20 | 201 | Homo sapiens | Mutation(s): 0 Gene Names: ARHGAP20, KIAA1391 | 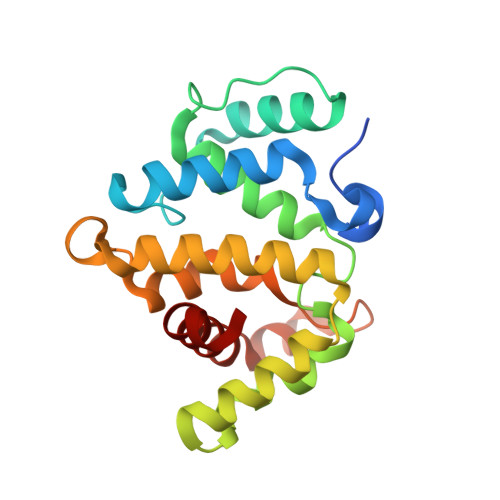 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9P2F6 (Homo sapiens) Explore Q9P2F6 Go to UniProtKB: Q9P2F6 | |||||
PHAROS: Q9P2F6 GTEx: ENSG00000137727 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9P2F6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GDP Query on GDP | C [auth A] | GUANOSINE-5'-DIPHOSPHATE C10 H15 N5 O11 P2 QGWNDRXFNXRZMB-UUOKFMHZSA-N |  | ||
| MGF Query on MGF | E [auth A] | TRIFLUOROMAGNESATE F3 Mg GJOMWUHGUQLOAC-UHFFFAOYSA-K |  | ||
| MG Query on MG | D [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 69.057 | α = 90 |
| b = 88.977 | β = 90 |
| c = 137.725 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| AMoRE | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| SERGUI | data collection |