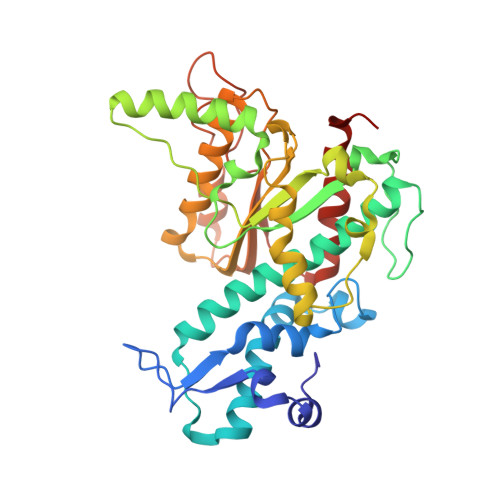
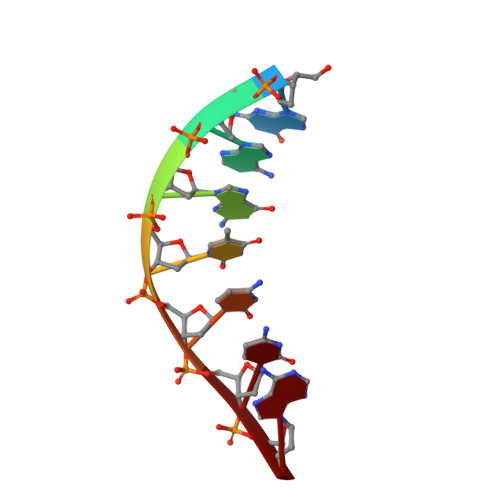
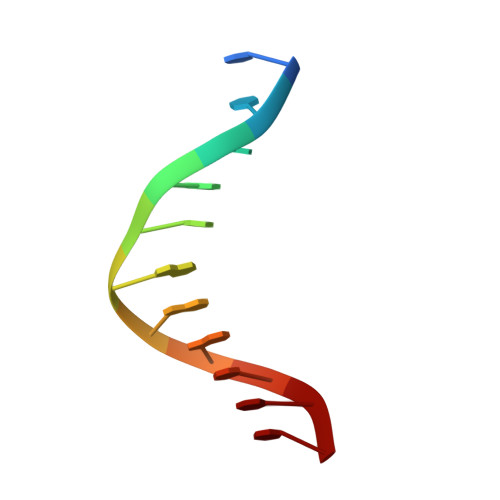
New clues in the allosteric activation of DNA cleavage by SgrAI: structures of SgrAI bound to cleaved primary-site DNA and uncleaved secondary-site DNA.
Little, E.J., Dunten, P.W., Bitinaite, J., Horton, N.C.(2011) Acta Crystallogr D Biol Crystallogr 67: 67-74
- PubMed: 21206063
- DOI: https://doi.org/10.1107/S0907444910047785
- Primary Citation of Related Structures:
3MQY, 3N78, 3N7B - PubMed Abstract:
SgrAI is a type II restriction endonuclease that cuts an unusually long recognition sequence and exhibits allosteric self-activation with expansion of DNA-sequence specificity. The three-dimensional crystal structures of SgrAI bound to cleaved primary-site DNA and Mg²(+) and bound to secondary-site DNA with either Mg²(+) or Ca²(+) are presented. All three structures show a conformation of enzyme and DNA similar to the previously determined dimeric structure of SgrAI bound to uncleaved primary-site DNA and Ca²(+) [Dunten et al. (2008), Nucleic Acids Res. 36, 5405-5416], with the exception of the cleaved bond and a slight shifting of the DNA in the SgrAI/cleaved primary-site DNA/Mg²(+) structure. In addition, a new metal ion binding site is located in one of the two active sites in this structure, which is consistent with proposals for the existence of a metal-ion site near the 3'-O leaving group.
- Department of Chemistry and Biochemistry, University of Arizona, Tucson, 85721, USA.
Organizational Affiliation: