Characterization of the self-palmitoylation activity of the transport protein particle component Bet3
Kummel, D., Walter, J., Heck, M., Heinemann, U., Veit, M.(2010) Cell Mol Life Sci 67: 2653-2664
- PubMed: 20372964
- DOI: https://doi.org/10.1007/s00018-010-0358-y
- Primary Citation of Related Structures:
3KXC - PubMed Abstract:
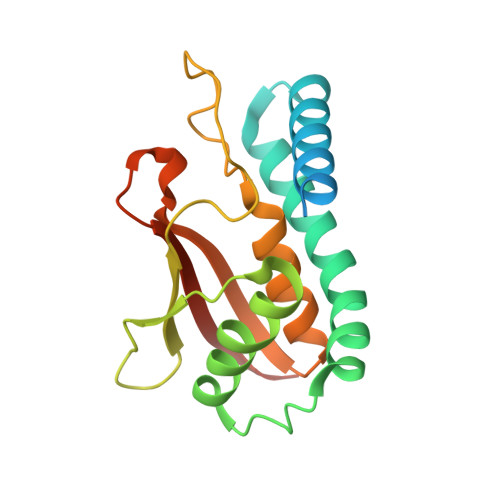
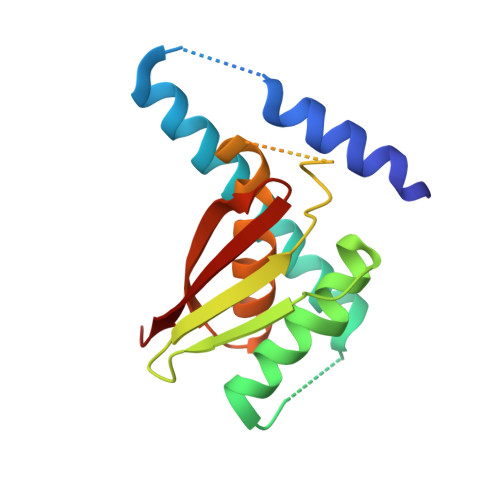
Bet3, a transport protein particle component involved in vesicular trafficking, contains a hydrophobic tunnel occupied by a fatty acid linked to cysteine 68. We reported that Bet3 has a unique self-palmitoylating activity. Here we show that mutation of arginine 67 reduced self-palmitoylation of Bet3, but the effect was compensated by increasing the pH. Thus, arginine helps to deprotonate cysteine such that it could function as a nucleophile in the acylation reaction which is supported by the structural analysis of non-acylated Bet3. Using fluorescence spectroscopy we show that long-chain acyl-CoAs bind with micromolar affinity to Bet3, whereas shorter-chain acyl-CoAs do not interact. Mutants with a deleted acylation site or a blocked tunnel bind to Pal-CoA, only the latter with slightly reduced affinity. Bet3 contains three binding sites for Pal-CoA, but their number was reduced to two in the mutant with an obstructed tunnel, indicating that Bet3 contains binding sites on its surface.
- Max Delbrück Center for Molecular Medicine, Robert-Rössle-Strasse 10, Berlin, Germany.
Organizational Affiliation: