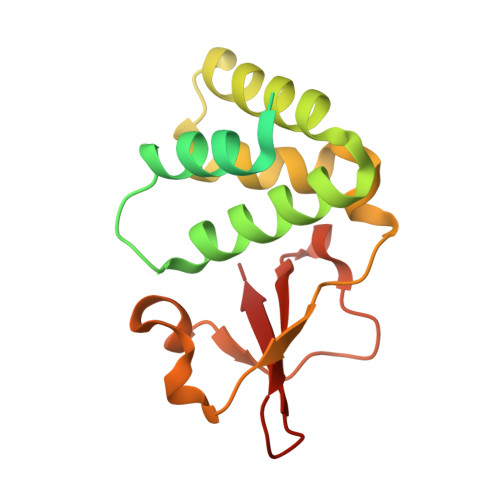
Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Kimberlin, C.R., Bornholdt, Z.A., Li, S., Woods, V.L., Macrae, I.J., Saphire, E.O.(2009) Proc Natl Acad Sci U S A 107: 314-319
- PubMed: 20018665
- DOI: https://doi.org/10.1073/pnas.0910547107
- Primary Citation of Related Structures:
3KS4, 3KS8 - PubMed Abstract:
Ebolavirus causes a severe hemorrhagic fever and is divided into five distinct species, of which Reston ebolavirus is uniquely nonpathogenic to humans. Disease caused by ebolavirus is marked by early immunosuppression of innate immune signaling events, involving silencing and sequestration of double-stranded RNA (dsRNA) by the viral protein VP35. Here we present unbound and dsRNA-bound crystal structures of the dsRNA-binding domain of Reston ebolavirus VP35. The structures show that VP35 forms an unusual, asymmetric dimer on dsRNA binding, with each of the monomers binding dsRNA in a different way: one binds the backbone whereas the other caps the terminus. Additional SAXS, DXMS, and dsRNA-binding experiments presented here support a model of cooperative dsRNA recognition in which binding of the first monomer assists binding of the next monomer of the oligomeric VP35 protein. This work illustrates how ebolavirus VP35 could mask key recognition sites of molecules such as RIG-I, MDA-5, and Dicer to silence viral dsRNA in infection.
- Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: