X-ray crystal structure of C-terminal domain of Peptide Chain Release Factor from Methanosarcina mazei.
Osipiuk, J., Rakowski, E., Freeman, L., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptide chain release factor subunit 1 | 166 | Methanosarcina mazei | Mutation(s): 0 Gene Names: MM_1347, prf1 | 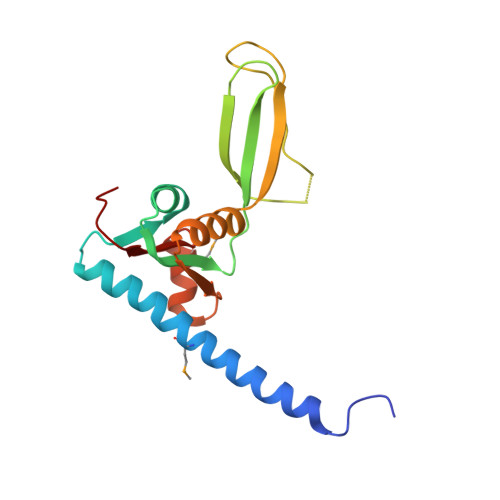 | |
UniProt | |||||
Find proteins for Q8PX75 (Methanosarcina mazei (strain ATCC BAA-159 / DSM 3647 / Goe1 / Go1 / JCM 11833 / OCM 88)) Explore Q8PX75 Go to UniProtKB: Q8PX75 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8PX75 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | C [auth A], D [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 116.124 | α = 90 |
| b = 116.124 | β = 90 |
| c = 71.454 | γ = 120 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| SBC-Collect | data collection |
| HKL-3000 | data reduction |
| SHELXD | phasing |
| MLPHARE | phasing |
| DM | phasing |
| SOLVE | phasing |
| RESOLVE | phasing |
| HKL-3000 | phasing |