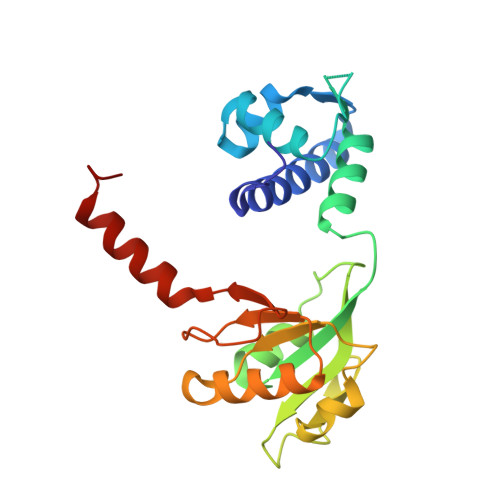
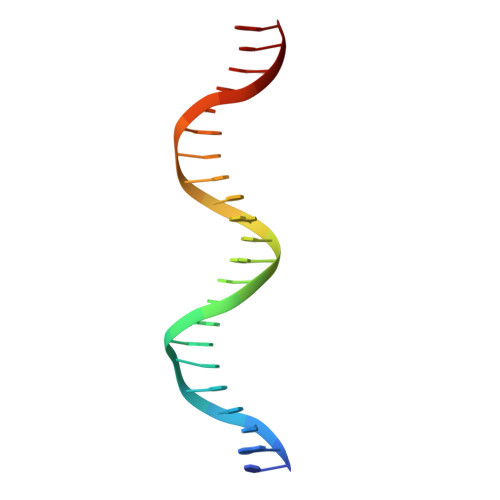
Structural basis for NADH/NAD+ redox sensing by a Rex family repressor.
McLaughlin, K.J., Strain-Damerell, C.M., Xie, K., Brekasis, D., Soares, A.S., Paget, M.S., Kielkopf, C.L.(2010) Mol Cell 38: 563-575
- PubMed: 20513431
- DOI: https://doi.org/10.1016/j.molcel.2010.05.006
- Primary Citation of Related Structures:
3IKT, 3IKV, 3IL2 - PubMed Abstract:
Nicotinamide adenine dinucleotides have emerged as key signals of the cellular redox state. Yet the structural basis for allosteric gene regulation by the ratio of reduced NADH to oxidized NAD(+) is poorly understood. A key sensor among Gram-positive bacteria, Rex represses alternative respiratory gene expression until a limited oxygen supply elevates the intracellular NADH:NAD(+) ratio. Here we investigate the molecular mechanism for NADH/NAD(+) sensing among Rex family members by determining structures of Thermus aquaticus Rex bound to (1) NAD(+), (2) DNA operator, and (3) without ligand. Comparison with the Rex/NADH complex reveals that NADH releases Rex from the DNA site following a 40 degrees closure between the dimeric subunits. Complementary site-directed mutagenesis experiments implicate highly conserved residues in NAD-responsive DNA-binding activity. These rare views of a redox sensor in action establish a means for slight differences in the nicotinamide charge, pucker, and orientation to signal the redox state of the cell.
- Department of Biochemistry and Biophysics, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA.
Organizational Affiliation: