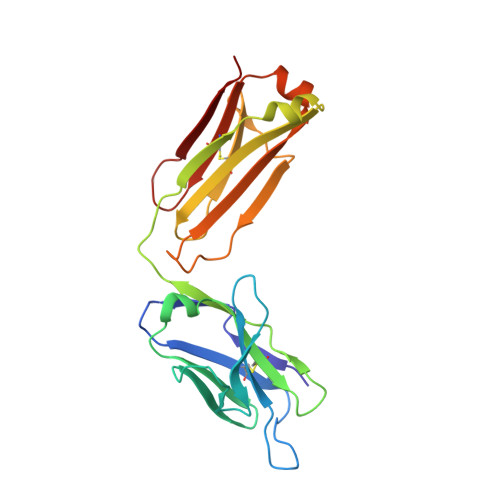
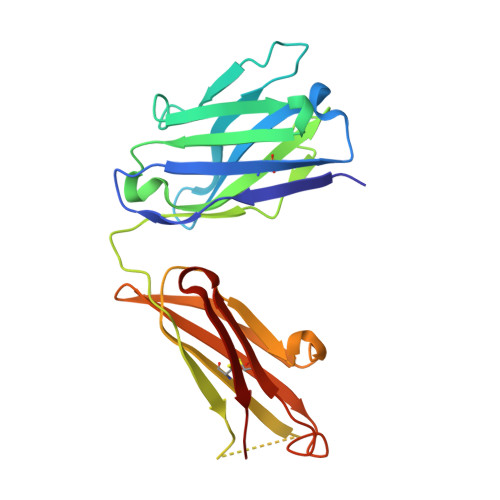
Antibody recognition of a unique tumor-specific glycopeptide antigen.
Brooks, C.L., Schietinger, A., Borisova, S.N., Kufer, P., Okon, M., Hirama, T., Mackenzie, C.R., Wang, L.X., Schreiber, H., Evans, S.V.(2010) Proc Natl Acad Sci U S A 107: 10056-10061
- PubMed: 20479270
- DOI: https://doi.org/10.1073/pnas.0915176107
- Primary Citation of Related Structures:
3IET, 3IF1 - PubMed Abstract:
Aberrant glycosylation and the overexpression of certain carbohydrate moieties is a consistent feature of cancers, and tumor-associated oligosaccharides are actively investigated as targets for immunotherapy. One of the most common aberrations in glycosylation patterns is the presentation of a single O-linked N-acetylgalactosamine on a threonine or serine residue known as the "Tn antigen." Whereas the ubiquitous nature of Tn antigens on cancers has made them a natural focus of vaccine research, such carbohydrate moieties are not always tumor-specific and have been observed on embryonic and nonmalignant adult tissue. Here we report the structural basis of binding of a complex of a monoclonal antibody (237mAb) with a truly tumor-specific glycopeptide containing the Tn antigen. In contrast to glycopeptide-specific antibodies in complex with simple peptides, 237mAb does not recognize a conformational epitope induced in the peptide by sugar substitution. Instead, 237mAb uses a pocket coded by germ-line genes to completely envelope the carbohydrate moiety itself while interacting with the peptide moiety in a shallow groove. Thus, 237mAb achieves its striking tumor specificity, with no observed physiological cross-reactivity to the unglycosylated peptide or the free glycan, by a combination of multiple weak but specific interactions to both the peptide and to the glycan portions of the antigen.
- Department of Biochemistry and Microbiology, University of Victoria, Victoria, BC, Canada V8P 3P6.
Organizational Affiliation: