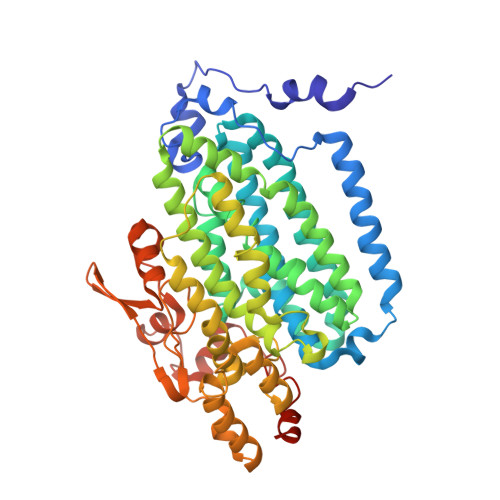
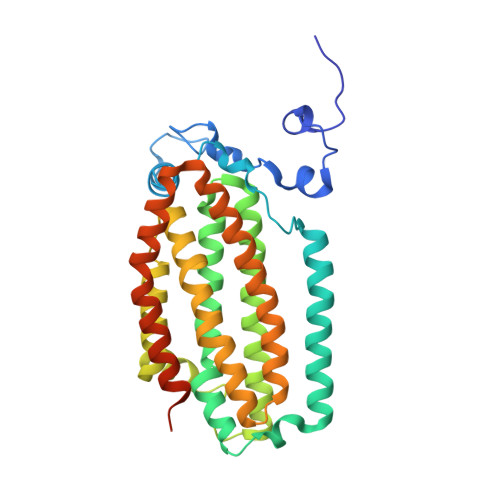
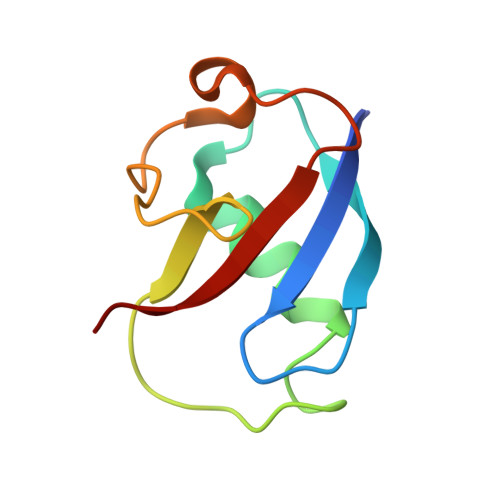
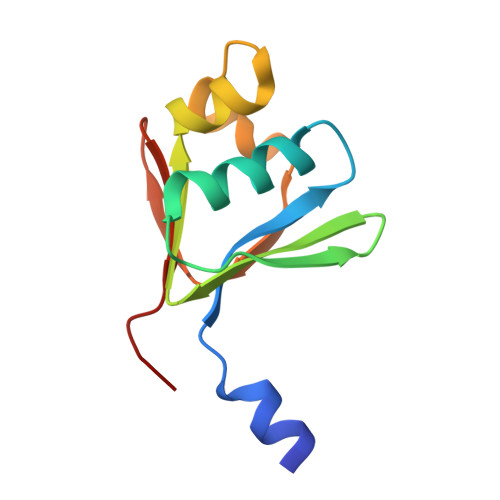
Crystallographic and catalytic studies of the peroxide-shunt reaction in a diiron hydroxylase.
Bailey, L.J., Fox, B.G.(2009) Biochemistry 48: 8932-8939
- PubMed: 19705873
- DOI: https://doi.org/10.1021/bi901150a
- Primary Citation of Related Structures:
3I5J, 3I63 - PubMed Abstract:
A diiron hydroxylase reaction typically begins by combination of O2 with a diferrous center to form reactive intermediates capable of hydrocarbon hydroxylation. In this natural cycle, reducing equivalents are provided by specific interactions with electron transfer proteins. The biological process can be bypassed by combining H2O2 with a diferric center, i.e., peroxide-shunt catalysis. Here we show that toluene 4-monooxygenase has a peroxide-shunt reaction that is approximately 600-fold slower than catalysis driven by biological electron transfer. However, the toluene 4-monooxygenase hydroxylase-effector protein complex was stable in the presence of 300 mM H2O2, suggesting overall benign effects of the exogenous oxidant on active site structure and function. The X-ray structure of the toluene 4-monooxygenase hydroxylase-effector protein complex determined from crystals soaked in H2O2 revealed a bound diatomic molecule, assigned to a cis-mu-1,2-peroxo bridge. This peroxo species resides in an active site position adjacent to the hydrogen-bonding substructure established by effector protein binding and faces into the distal cavity where substrate must bind during regiospecific aromatic ring hydroxylation catalysis. These results provide a new structural benchmark for how activated intermediates may be formed and dispatched during diiron hydroxylase catalysis.
- Department of Biochemistry, College of Agricultural and Life Sciences, University of Wisconsin, Madison, Wisconsin 53706-1544, USA.
Organizational Affiliation: