Hydrogen bonds at the protein-inhibitor interface in the HIV-1 protease / inhibitors complexes probed by total chemical synthesis and X-ray crystallography
Torbeev, V.Y., Kent, S.B.H.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HIV-1 protease | 99 | N/A | Mutation(s): 0 EC: 3.4.23.16 | 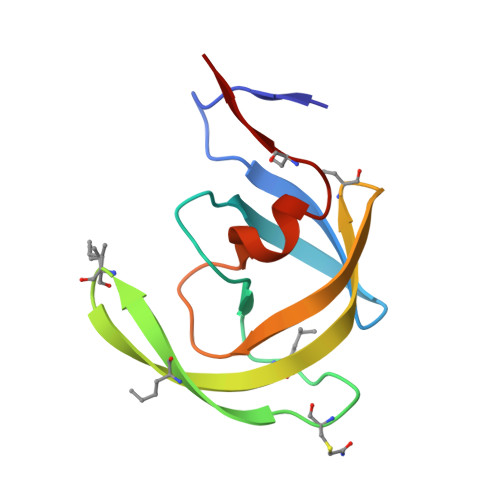 | |
UniProt | |||||
Find proteins for P03369 (Human immunodeficiency virus type 1 group M subtype B (isolate ARV2/SF2)) Explore P03369 Go to UniProtKB: P03369 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03369 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 2NC Query on 2NC | C [auth B] | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide C35 H68 N11 O8 MQPXOVRKKPPKFZ-QYKDHROSSA-O |  | ||
| Modified Residues 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| ABA Query on ABA | A, B | L-PEPTIDE LINKING | C4 H9 N O2 |  | ALA |
| IIL Query on IIL | A, B | L-PEPTIDE LINKING | C6 H13 N O2 |  | ILE |
| NLE Query on NLE | A, B | L-PEPTIDE LINKING | C6 H13 N O2 |  | LEU |
| YCM Query on YCM | A, B | L-PEPTIDE LINKING | C5 H10 N2 O3 S |  | CYS |
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_000398 (2NC) Query on PRD_000398 | C [auth B] | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide | Peptide-like / Inhibitor |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 51.139 | α = 90 |
| b = 58.084 | β = 90 |
| c = 61.525 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |