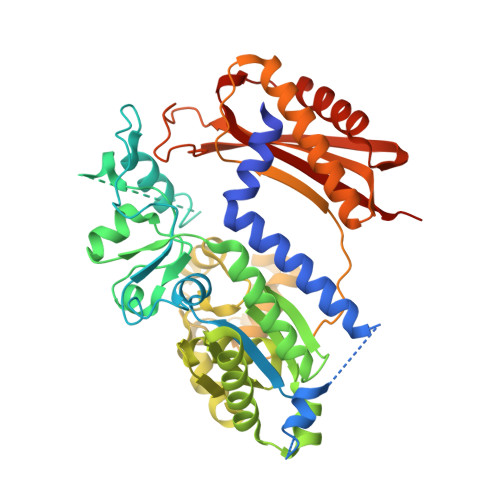
Structure of the human Rev1-DNA-dNTP ternary complex.
Swan, M.K., Johnson, R.E., Prakash, L., Prakash, S., Aggarwal, A.K.(2009) J Mol Biology 390: 699-709
- PubMed: 19464298
- DOI: https://doi.org/10.1016/j.jmb.2009.05.026
- Primary Citation of Related Structures:
3GQC - PubMed Abstract:
Y-family DNA polymerases have proven to be remarkably diverse in their functions and in strategies for replicating through DNA lesions. The structure of yeast Rev1 ternary complex has revealed the most radical replication strategy, where the polymerase itself dictates the identity of the incoming nucleotide, as well as the identity of the templating base. We show here that many of the key elements of this highly unusual strategy are conserved between yeast and human Rev1, including the eviction of template G from the DNA helix and the pairing of incoming deoxycytidine 5'-triphosphate with a surrogate arginine residue. We also show that the catalytic core of human Rev1 is uniquely augmented by two large inserts, I1 and I2, wherein I1 extends >20 A away from the active site and may serve as a platform for protein-protein interactions specific for Rev1's role in translesion DNA synthesis in human cells, and I2 acts as a "flap" on the hydrophobic pocket accommodating template G. We suggest that these novel structural features are important for providing human Rev1 greater latitude in promoting efficient and error-free translesion DNA synthesis through the diverse array of bulky and potentially carcinogenic N(2)-deoxyguanosine DNA adducts in human cells.
- Department of Structural and Chemical Biology, Mount Sinai School of Medicine, New York, NY 10029, USA.
Organizational Affiliation: