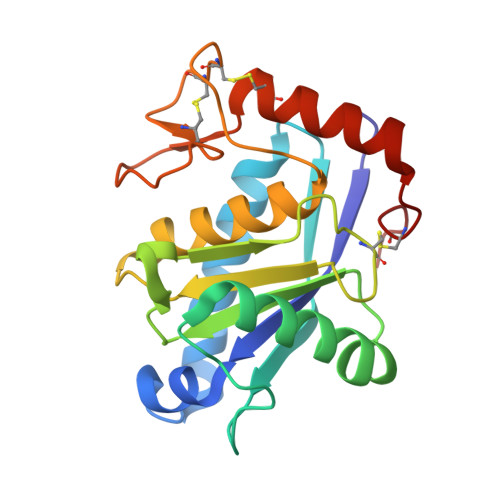
Structural studies of BmooMPalpha-I, a non-hemorrhagic metalloproteinase from Bothrops moojeni venom.
Akao, P.K., Tonoli, C.C., Navarro, M.S., Cintra, A.C., Neto, J.R., Arni, R.K., Murakami, M.T.(2010) Toxicon 55: 361-368
- PubMed: 19706302
- DOI: https://doi.org/10.1016/j.toxicon.2009.08.013
- Primary Citation of Related Structures:
3GBO - PubMed Abstract:
Hemostatically active snake venom metalloproteinases (SVMPs) perturb the blood coagulation cascade at specific points and due to their potential application as thrombolytic agents, the fibrin(ogen)olytic non-hemorrhagic SVMPs have been employed as biochemical tools in coagulation research and diagnosis. Structural studies complemented by the design of metalloproteinase inhibitors have been instrumental in understanding their stereo specificity and action mechanism. We present here, details of the crystal structure of BmooMPalpha-I, a 22.6 kDa non-hemorrhagic P-I class SVMP isolated from Bothrops moojeni venom, determined at 1.76 A resolution. In this structure, the catalytic zinc ion displays an unusual octahedral coordination formed by the three canonical histidines (His(142), His(146) and His(152)) and additionally, by three solvent molecules. Comparative sequence and structural studies indicate that the motif comprising amino acid segments 153-164 and 167-176 adjacent to the methionine-turn is a salient feature that differentiates both non and hemorrhagic P-I class SVMPs and could directly be involved in the development of the hemorrhagic activity.
- Center for Structural Molecular Biology, Brazilian Synchrotron Light Laboratory, Campinas, SP 13083-970, Brazil.
Organizational Affiliation: