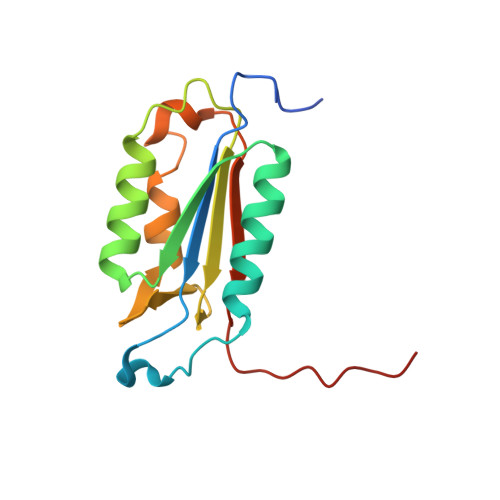
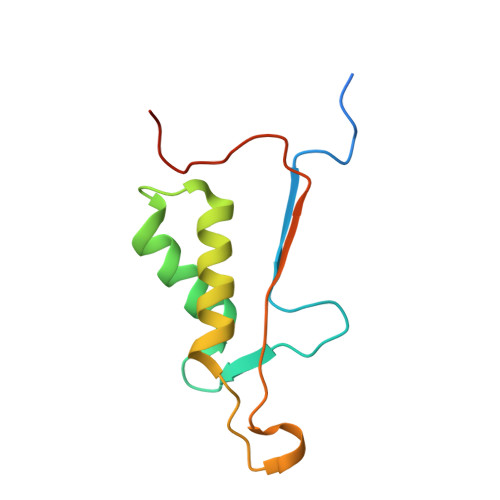
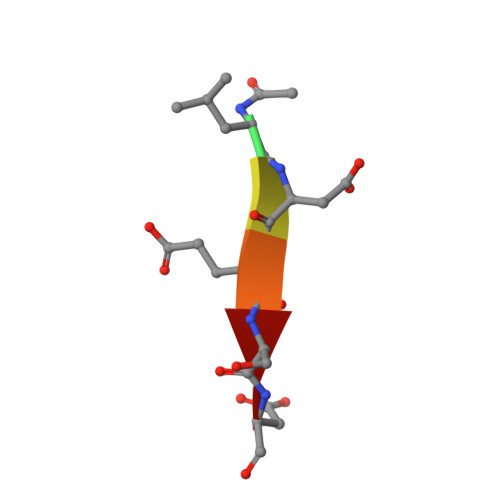
Structural basis for executioner caspase recognition of P5 position in substrates.
Fu, G., Chumanevich, A.A., Agniswamy, J., Fang, B., Harrison, R.W., Weber, I.T.(2008) Apoptosis 13: 1291-1302
- PubMed: 18780184
- DOI: https://doi.org/10.1007/s10495-008-0259-9
- Primary Citation of Related Structures:
3EDQ, 3EDR - PubMed Abstract:
Caspase-3, -6 and -7 cleave many proteins at specific sites to induce apoptosis. Their recognition of the P5 position in substrates has been investigated by kinetics, modeling and crystallography. Caspase-3 and -6 recognize P5 in pentapeptides as shown by enzyme activity data and interactions observed in the crystal structure of caspase-3/LDESD and in a model for caspase-6. In caspase-3 the P5 main-chain was anchored by interactions with Ser209 in loop-3 and the P5 Leu side-chain interacted with Phe250 and Phe252 in loop-4 consistent with 50% increased hydrolysis of LDEVD relative to DEVD. Caspase-6 formed similar interactions and showed a preference for polar P5 in QDEVD likely due to interactions with polar Lys265 and hydrophobic Phe263 in loop-4. Caspase-7 exhibited no preference for P5 residue in agreement with the absence of P5 interactions in the caspase-7/LDESD crystal structure. Initiator caspase-8, with Pro in the P5-anchoring position and no loop-4, had only 20% activity on tested pentapeptides relative to DEVD. Therefore, caspases-3 and -6 bind P5 using critical loop-3 anchoring Ser/Thr and loop-4 side-chain interactions, while caspase-7 and -8 lack P5-binding residues.
- Department of Biology, Molecular Basis of Disease Program, Georgia State University, Atlanta, GA 30303, USA.
Organizational Affiliation: