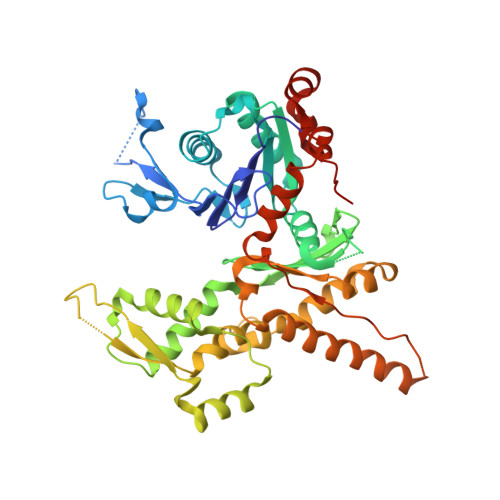
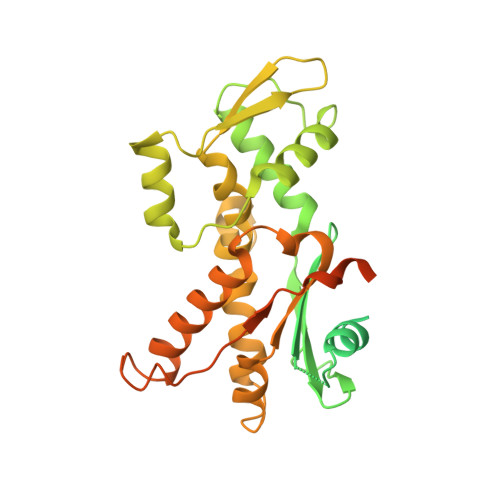
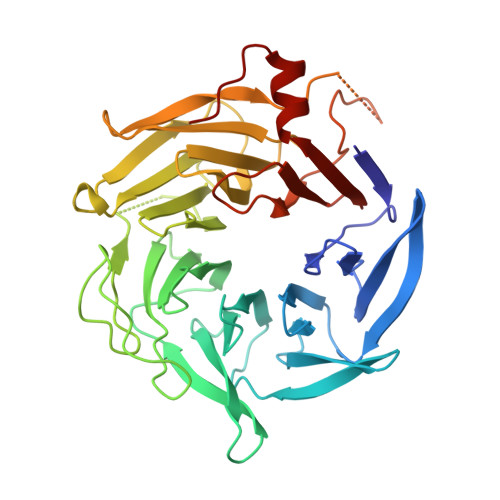
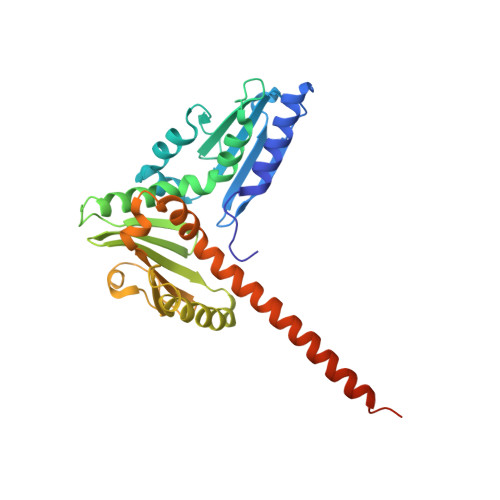
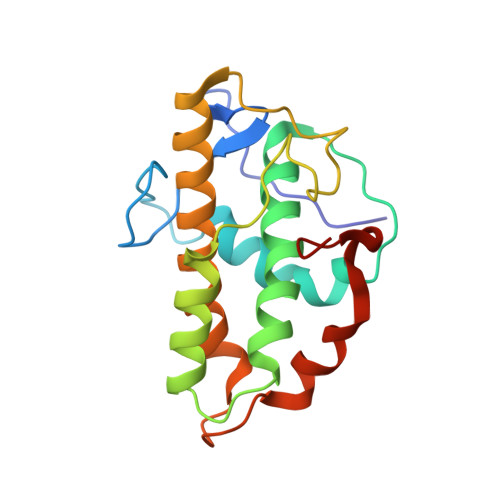
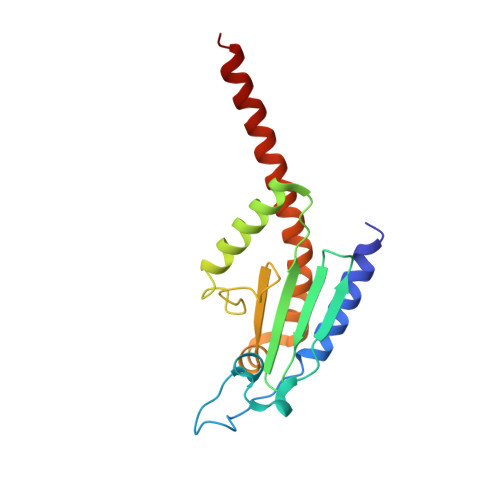
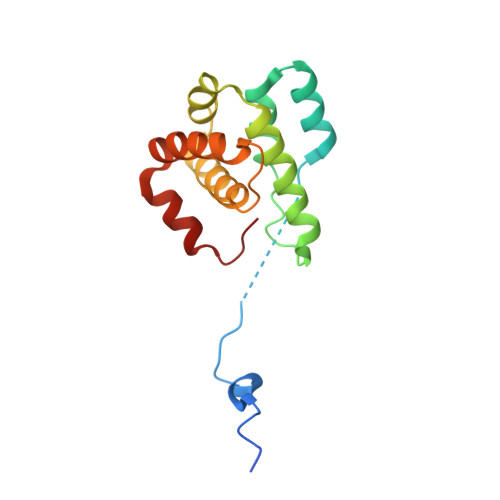
Characterization of two classes of small molecule inhibitors of Arp2/3 complex
Nolen, B.J., Tomasevic, N., Russell, A., Pierce, D.W., Jia, Z., McCormick, C.D., Hartman, J., Sakowicz, R., Pollard, T.D.(2009) Nature 460: 1031-1034
- PubMed: 19648907
- DOI: https://doi.org/10.1038/nature08231
- Primary Citation of Related Structures:
3DXK, 3DXM - PubMed Abstract:
Polymerization of actin filaments directed by the actin-related protein (Arp)2/3 complex supports many types of cellular movements. However, questions remain regarding the relative contributions of Arp2/3 complex versus other mechanisms of actin filament nucleation to processes such as path finding by neuronal growth cones; this is because of the lack of simple methods to inhibit Arp2/3 complex reversibly in living cells. Here we describe two classes of small molecules that bind to different sites on the Arp2/3 complex and inhibit its ability to nucleate actin filaments. CK-0944636 binds between Arp2 and Arp3, where it appears to block movement of Arp2 and Arp3 into their active conformation. CK-0993548 inserts into the hydrophobic core of Arp3 and alters its conformation. Both classes of compounds inhibit formation of actin filament comet tails by Listeria and podosomes by monocytes. Two inhibitors with different mechanisms of action provide a powerful approach for studying the Arp2/3 complex in living cells.
- Department of Molecular Cellular and Developmental Biology, Yale University, New Haven, Connecticut 06520, USA.
Organizational Affiliation: