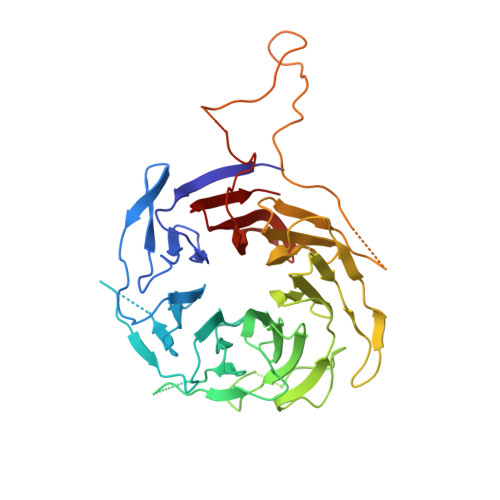
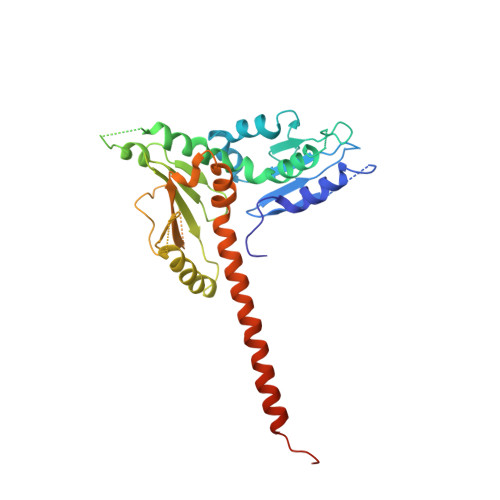
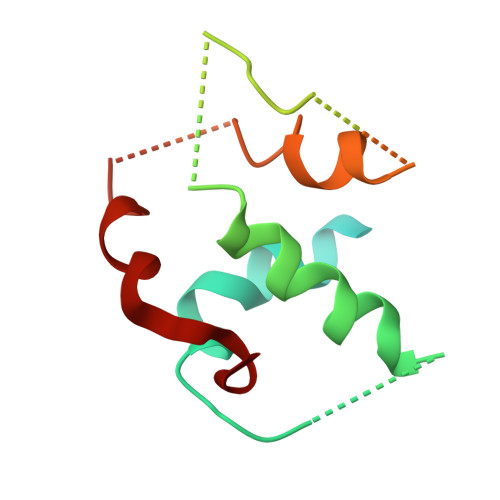
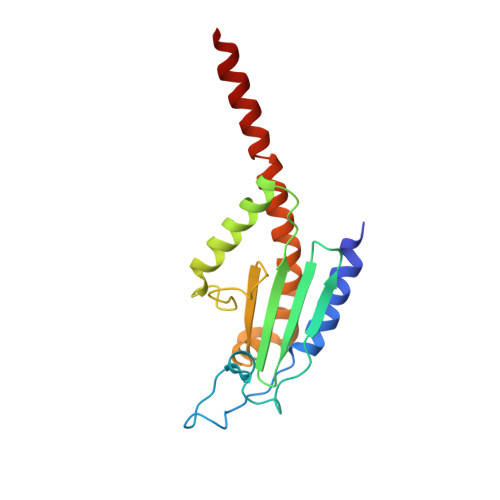
Structure and biochemical properties of fission yeast arp2/3 complex lacking the arp2 subunit.
Nolen, B.J., Pollard, T.D.(2008) J Biological Chem 283: 26490-26498
- PubMed: 18640983
- DOI: https://doi.org/10.1074/jbc.M802607200
- Primary Citation of Related Structures:
3DWL - PubMed Abstract:
Arp2/3 (actin-related protein 2/3) complex is a seven-subunit complex that nucleates branched actin filaments in response to cellular signals. Nucleation-promoting factors such as WASp/Scar family proteins activate the complex by facilitating the activating conformational change and recruiting the first actin monomer for the daughter branch. Here we address the role of the Arp2 subunit in the function of Arp2/3 complex by isolating a version of the complex lacking Arp2 (Arp2Delta Arp2/3 complex) from fission yeast. An x-ray crystal structure of the DeltaArp2 Arp2/3 complex showed that the rest of the complex is unperturbed by the loss of Arp2. However, the Arp2Delta Arp2/3 complex was inactive in actin nucleation assays, indicating that Arp2 is essential to form a branch. A fluorescence anisotropy assay showed that Arp2 does not contribute to the affinity of the complex for Wsp1-VCA, a Schizosaccharomyces pombe nucleation-promoting factor protein. Fluorescence resonance energy transfer experiments showed that the loss of Arp2 does not prevent VCA from recruiting an actin monomer to the complex. Truncation of the N terminus of ARPC5, the smallest subunit in the complex, increased the yield of Arp2Delta Arp2/3 complex during purification but did not compromise nucleation activity of the full Arp2/3 complex.
- Departments of Molecular, Cellular and Developmental Biology, Yale University, New Haven, Connecticut 06520-8103, USA. bradley.nolen@yale.edu
Organizational Affiliation: