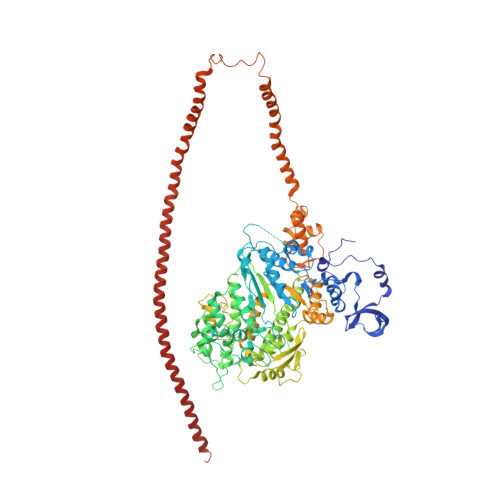
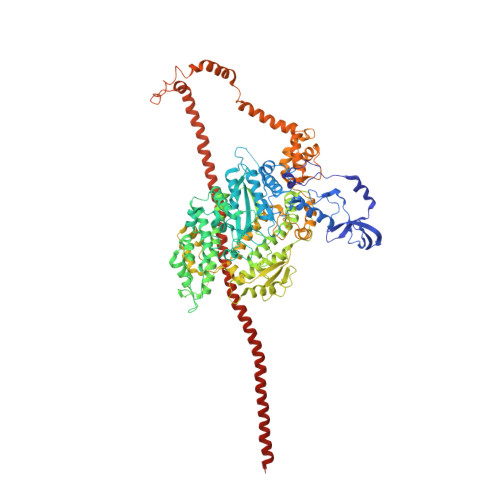
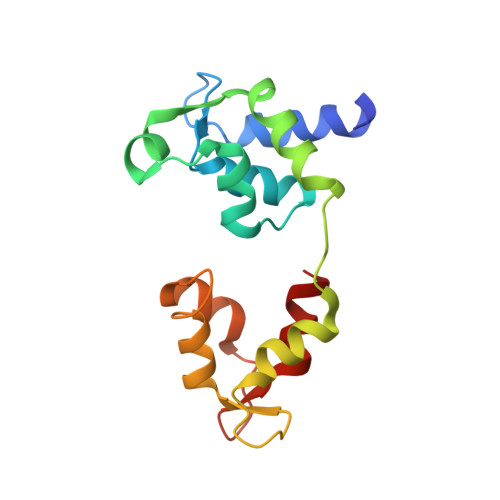
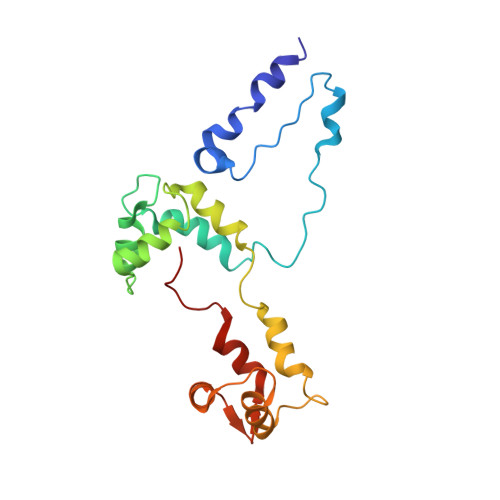
Three-Dimensional Reconstruction of Tarantula Myosin Filaments Suggests How Phosphorylation May Regulate Myosin Activity
Alamo, L., Wriggers, W., Pinto, A., Bartoli, F., Salazar, L., Zhao, F.Q., Craig, R., Padron, R.(2008) J Mol Biology 384: 780-797
- PubMed: 18951904
- DOI: https://doi.org/10.1016/j.jmb.2008.10.013
- Primary Citation of Related Structures:
3DTP - PubMed Abstract:
Muscle contraction involves the interaction of the myosin heads of the thick filaments with actin subunits of the thin filaments. Relaxation occurs when this interaction is blocked by molecular switches on these filaments. In many muscles, myosin-linked regulation involves phosphorylation of the myosin regulatory light chains (RLCs). Electron microscopy of vertebrate smooth muscle myosin molecules (regulated by phosphorylation) has provided insight into the relaxed structure, revealing that myosin is switched off by intramolecular interactions between its two heads, the free head and the blocked head. Three-dimensional reconstruction of frozen-hydrated specimens revealed that this asymmetric head interaction is also present in native thick filaments of tarantula striated muscle. Our goal in this study was to elucidate the structural features of the tarantula filament involved in phosphorylation-based regulation. A new reconstruction revealed intra- and intermolecular myosin interactions in addition to those seen previously. To help interpret the interactions, we sequenced the tarantula RLC and fitted an atomic model of the myosin head that included the predicted RLC atomic structure and an S2 (subfragment 2) crystal structure to the reconstruction. The fitting suggests one intramolecular interaction, between the cardiomyopathy loop of the free head and its own S2, and two intermolecular interactions, between the cardiac loop of the free head and the essential light chain of the blocked head and between the Leu305-Gln327 interaction loop of the free head and the N-terminal fragment of the RLC of the blocked head. These interactions, added to those previously described, would help switch off the thick filament. Molecular dynamics simulations suggest how phosphorylation could increase the helical content of the RLC N-terminus, weakening these interactions, thus releasing both heads and activating the thick filament.
- Departamento de Biología Estructural, Instituto Venezolano de Investigaciones Científicas, Caracas, Venezuela.
Organizational Affiliation: