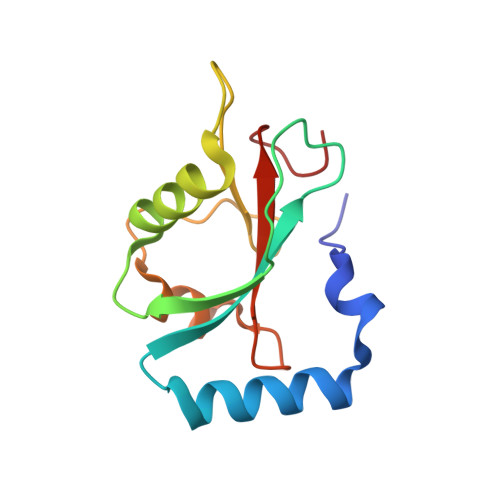
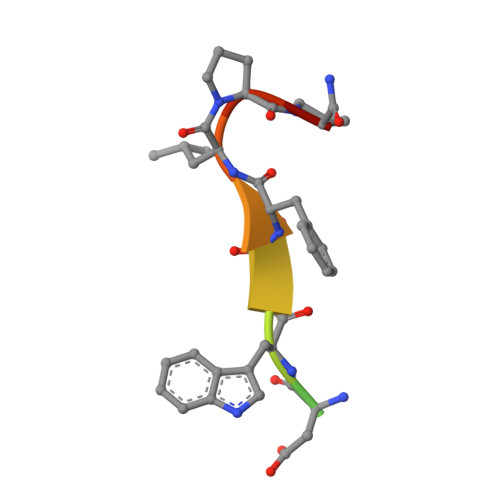
Structural framework of the GABARAP-calreticulin interface - implications for substrate binding to endoplasmic reticulum chaperones.
Thielmann, Y., Weiergraber, O.H., Mohrluder, J., Willbold, D.(2009) FEBS J 276: 1140-1152
- PubMed: 19154346
- DOI: https://doi.org/10.1111/j.1742-4658.2008.06857.x
- Primary Citation of Related Structures:
3DOW - PubMed Abstract:
The 4-aminobutyrate type A receptor-associated protein (GABARAP) is a versatile adaptor protein that plays an important role in intracellular vesicle trafficking, particularly in neuronal cells. We have investigated the structural determinants underlying the interaction of GABARAP with calreticulin using spectroscopic and crystallographic techniques. Specifically, we present the crystal structure of GABARAP in complex with its major binding epitope on the chaperone. Molecular modeling of a complex containing full-length calreticulin suggests a novel mode of substrate interaction, which may have functional implications for the calreticulin/calnexin family in general.
- Institut für Neurowissenschaften und Biophysik, Molekulare Biophysik, Forschungszentrum Jülich, Germany.
Organizational Affiliation: