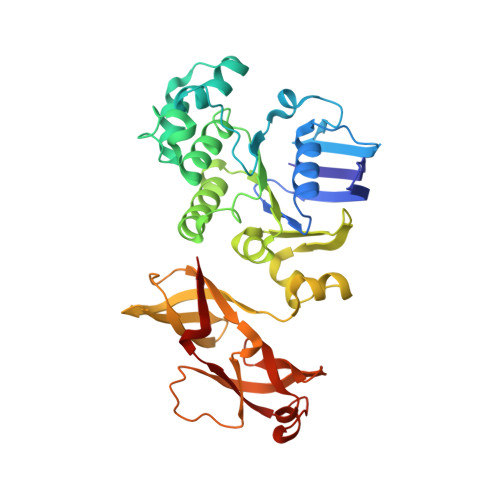
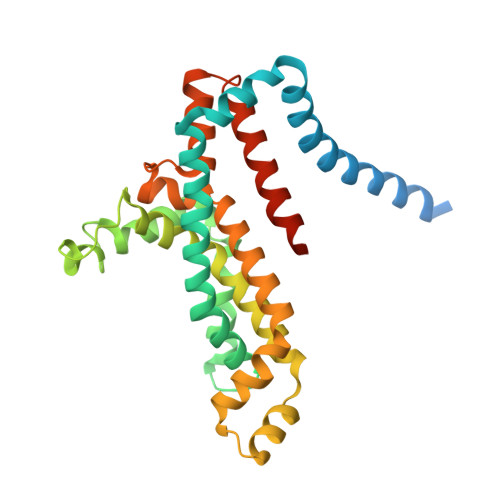
Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter.
Gerber, S., Comellas-Bigler, M., Goetz, B.A., Locher, K.P.(2008) Science 321: 246-250
- PubMed: 18511655
- DOI: https://doi.org/10.1126/science.1156213
- Primary Citation of Related Structures:
3D31 - PubMed Abstract:
Transport across cellular membranes is an essential process that is catalyzed by diverse membrane transport proteins. The turnover rates of certain transporters are inhibited by their substrates in a process termed trans-inhibition, whose structural basis is poorly understood. We present the crystal structure of a molybdate/tungstate ABC transporter (ModBC) from Methanosarcina acetivorans in a trans-inhibited state. The regulatory domains of the nucleotide-binding subunits are in close contact and provide two oxyanion binding pockets at the shared interface. By specifically binding to these pockets, molybdate or tungstate prevent adenosine triphosphatase activity and lock the transporter in an inward-facing conformation, with the catalytic motifs of the nucleotide-binding domains separated. This allosteric effect prevents the transporter from switching between the inward-facing and the outward-facing states, thus interfering with the alternating access and release mechanism.
- Institute of Molecular Biology and Biophysics, ETH Zürich, HPK D14.3, 8093 Zürich, Switzerland.
Organizational Affiliation: