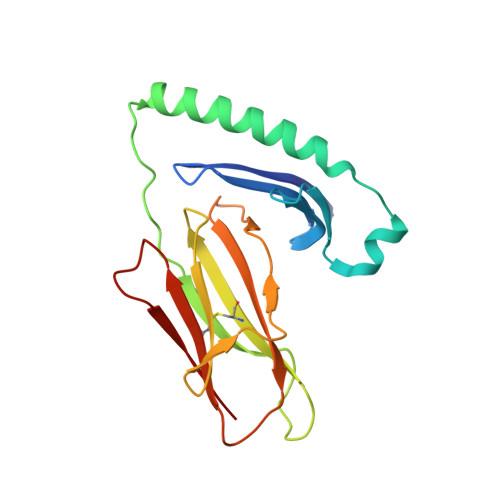
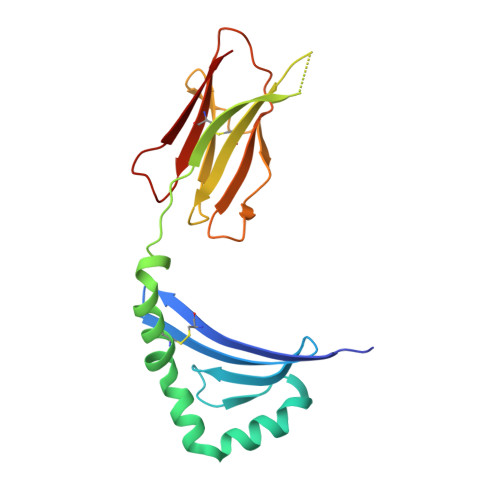
The structure of HLA-DR52c: comparison to other HLA-DRB3 alleles.
Dai, S., Crawford, F., Marrack, P., Kappler, J.W.(2008) Proc Natl Acad Sci U S A 105: 11893-11897
- PubMed: 18697946
- DOI: https://doi.org/10.1073/pnas.0805810105
- Primary Citation of Related Structures:
3C5J - PubMed Abstract:
Class II major histocompatibility complex (MHCII) molecules present antigens to CD4(+) T cells. In addition to the most commonly studied human MHCII isotype, HLA-DR, whose beta chain is encoded by the HLA-DRB1 locus, several other isotypes that use the same alpha chain but have beta chains encoded by other genes. These other DR molecules also are expressed in antigen-presenting cells and are known to participate in peptide presentation to T cells and to be recognized as alloantigens by other T cells. Like some of the HLA-DRB1 alleles, several of these alternate DR molecules have been associated with specific autoimmune diseases and T cell hypersensitivity. Here we present the structure of an HLA-DR molecule (DR52c) containing one of these alternate beta chains (HLA-DRB3*0301) bound to a self-peptide derived from the Tu elongation factor. The molecule shares structurally conserved elements with other MHC class II molecules but has some unique features in the peptide-binding groove. Comparison of the three major HLA-DBR3 alleles (DR52a, b, and c) suggests that they were derived from one another by recombination events that scrambled the four major peptide-binding pockets at peptide positions 1, 4, 6, and 9 but left virtually no polymorphisms elsewhere in the molecules.
- Integrated Department of Immunology, Howard Hughes Medical Institute, National Jewish Medical and Research Center, Denver, CO 80206, USA.
Organizational Affiliation: