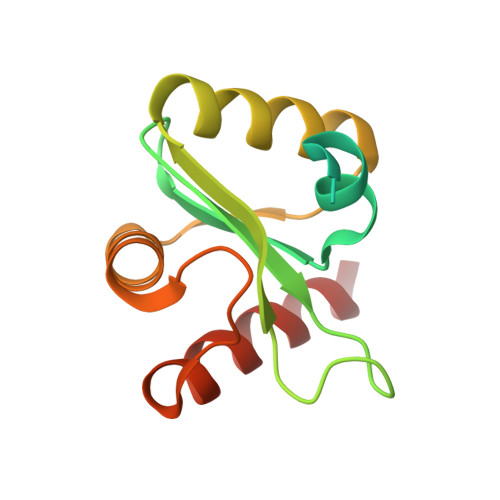
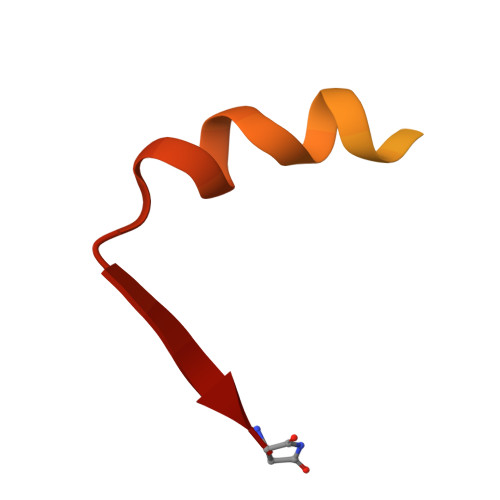
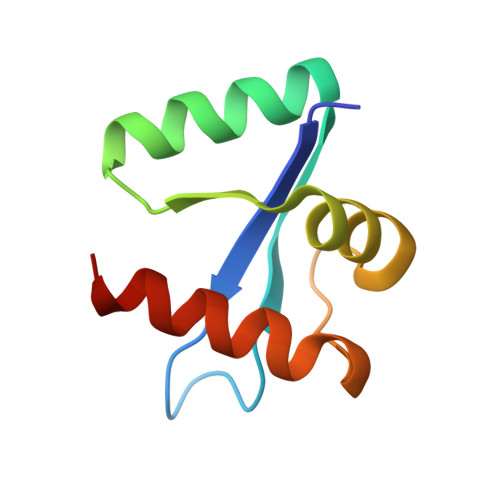
Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Zarivach, R., Deng, W., Vuckovic, M., Felise, H.B., Nguyen, H.V., Miller, S.I., Finlay, B.B., Strynadka, N.C.(2008) Nature 453: 124-127
- PubMed: 18451864
- DOI: https://doi.org/10.1038/nature06832
- Primary Citation of Related Structures:
3BZL, 3BZO, 3BZP, 3BZR, 3BZS, 3BZT, 3BZV, 3BZX, 3BZY, 3BZZ, 3C00, 3C01, 3C03 - PubMed Abstract:
During infection by Gram-negative pathogenic bacteria, the type III secretion system (T3SS) is assembled to allow for the direct transmission of bacterial virulence effectors into the host cell. The T3SS system is characterized by a series of prominent multi-component rings in the inner and outer bacterial membranes, as well as a translocation pore in the host cell membrane. These are all connected by a series of polymerized tubes that act as the direct conduit for the T3SS proteins to pass through to the host cell. During assembly of the T3SS, as well as the evolutionarily related flagellar apparatus, a post-translational cleavage event within the inner membrane proteins EscU/FlhB is required to promote a secretion-competent state. These proteins have long been proposed to act as a part of a molecular switch, which would regulate the appropriate chronological secretion of the various T3SS apparatus components during assembly and subsequently the transported virulence effectors. Here we show that a surface type II beta-turn in the Escherichia coli protein EscU undergoes auto-cleavage by a mechanism involving cyclization of a strictly conserved asparagine residue. Structural and in vivo analysis of point and deletion mutations illustrates the subtle conformational effects of auto-cleavage in modulating the molecular features of a highly conserved surface region of EscU, a potential point of interaction with other T3SS components at the inner membrane. In addition, this work provides new structural insight into the distinct conformational requirements for a large class of self-cleaving reactions involving asparagine cyclization.
- Department of Biochemistry and Molecular Biology, and the Center for Blood Research, University of British Columbia, 2350 Health Sciences Mall, Vancouver, British Columbia V6T 1Z3, Canada.
Organizational Affiliation: