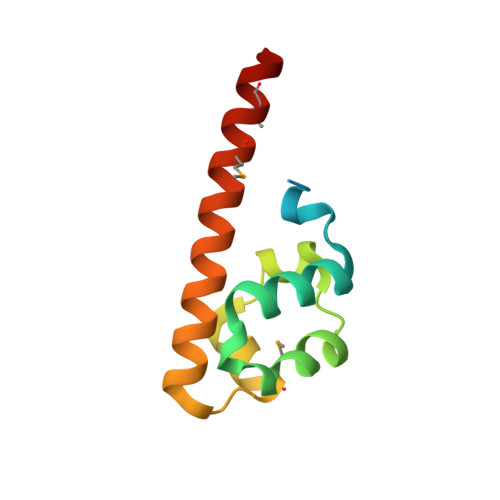
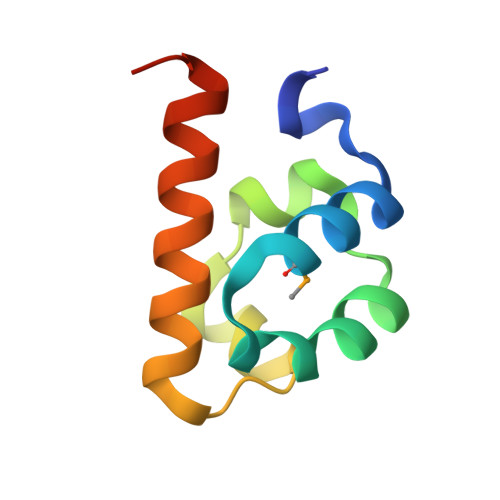
CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling.
Rajakulendran, T., Sahmi, M., Kurinov, I., Tyers, M., Therrien, M., Sicheri, F.(2008) Proc Natl Acad Sci U S A 105: 2836-2841
- PubMed: 18287031
- DOI: https://doi.org/10.1073/pnas.0709705105
- Primary Citation of Related Structures:
3BS5, 3BS7 - PubMed Abstract:
RAF kinase functions in the mitogen-activated protein kinase (MAPK) pathway to transmit growth signals to the downstream kinases MEK and ERK. Activation of RAF catalytic activity is facilitated by a regulatory complex comprising the proteins CNK (Connector enhancer of KSR), HYP (Hyphen), and KSR (Kinase Suppressor of Ras). The sterile alpha-motif (SAM) domain found in both CNK and HYP plays an essential role in complex formation. Here, we have determined the x-ray crystal structure of the SAM domain of CNK in complex with the SAM domain of HYP. The structure reveals a single-junction SAM domain dimer of 1:1 stoichiometry in which the binding mode is a variation of polymeric SAM domain interactions. Through in vitro and in vivo mutational analyses, we show that the specific mode of dimerization revealed by the crystal structure is essential for RAF signaling and facilitates the recruitment of KSR to form the CNK/HYP/KSR regulatory complex. We present two docking-site models to account for how SAM domain dimerization might influence the formation of a higher-order CNK/HYP/KSR complex.
- Centre for Systems Biology, Samuel Lunenfeld Research Institute, Toronto, ON, Canada.
Organizational Affiliation: