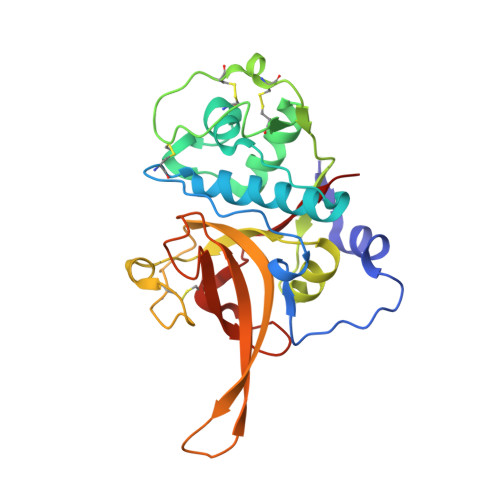
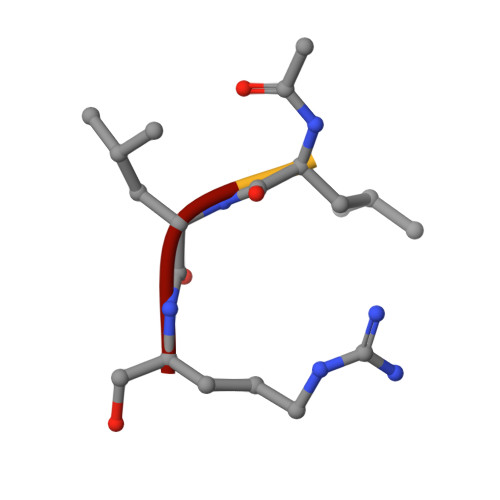
Structures of falcipain-2 and falcipain-3 bound to small molecule inhibitors: implications for substrate specificity.
Kerr, I.D., Lee, J.H., Pandey, K.C., Harrison, A., Sajid, M., Rosenthal, P.J., Brinen, L.S.(2009) J Med Chem 52: 852-857
- PubMed: 19128015
- DOI: https://doi.org/10.1021/jm8013663
- Primary Citation of Related Structures:
3BPF, 3BPM - PubMed Abstract:
Falcipain-2 and falcipain-3 are critical hemoglobinases of Plasmodium falciparum, the most virulent human malaria parasite. We have determined the 2.9 A crystal structure of falcipain-2 in complex with the epoxysuccinate E64 and the 2.5 A crystal structure of falcipain-3 in complex with the aldehyde leupeptin. These complexes represent the first crystal structures of plasmodial cysteine proteases with small molecule inhibitors and the first reported crystal structure of falcipain-3. Our structural analyses indicate that the relative shape and flexibility of the S2 pocket are affected by a number of discrete amino acid substitutions. The cumulative effect of subtle differences, including those at "gatekeeper" positions, may explain the observed kinetic differences between these two closely related enzymes.
- Department of Cellular and Molecular Pharmacology and Department of Pathology, University of California, San Francisco, California 94158, USA.
Organizational Affiliation: