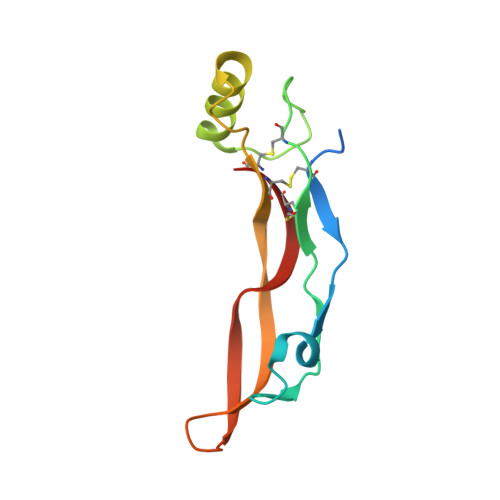
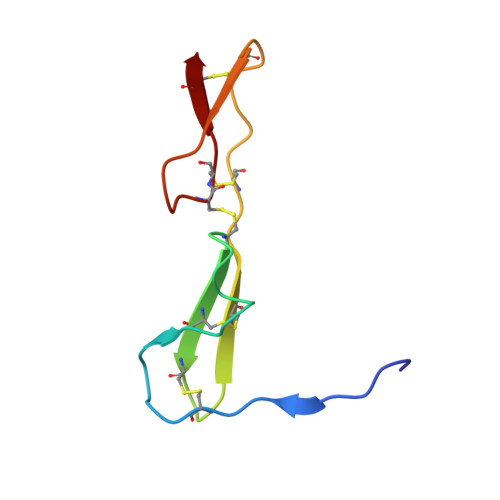
Crystal structure analysis reveals how the Chordin family member crossveinless 2 blocks BMP-2 receptor binding
Zhang, J.-L., Qiu, L.-Y., Kotzsch, A., Weidauer, S., Patterson, L., Hammerschmidt, M., Sebald, W., Mueller, T.D.(2008) Dev Cell 14: 739-750
- PubMed: 18477456
- DOI: https://doi.org/10.1016/j.devcel.2008.02.017
- Primary Citation of Related Structures:
3BK3 - PubMed Abstract:
Crossveinless 2 (CV-2) is an extracellular BMP modulator protein belonging to the Chordin family. During development it is expressed at sites of high BMP signaling and like Chordin CV-2 can either enhance or inhibit BMP activity. CV-2 binds to BMP-2 via its N-terminal Von Willebrand factor type C (VWC) domain 1. Here we report the structure of the complex between CV-2 VWC1 and BMP-2. The tripartite VWC1 binds BMP-2 only through a short N-terminal segment, called clip, and subdomain (SD) 1. Mutational analysis establishes that the clip segment and SD1 together create high-affinity BMP-2 binding. All four receptor-binding sites of BMP-2 are blocked in the complex, demonstrating that VWC1 acts as competitive inhibitor for all receptor types. In vivo experiments reveal that the BMP-enhancing (pro-BMP) activity of CV-2 is independent of BMP-2 binding by VWC1, showing that pro- and anti-BMP activities are structurally separated in CV-2.
- Department of Physiological Chemistry II, Biocenter, University of Wuerzburg, Am Hubland, 97074 Wuerzburg, Germany.
Organizational Affiliation: