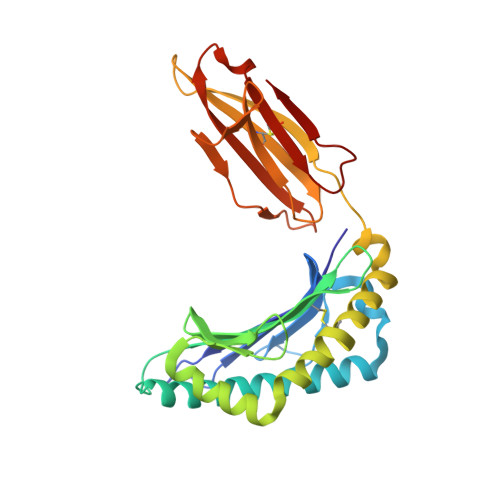
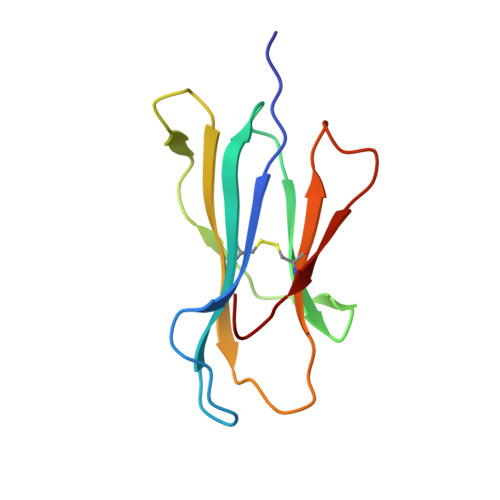
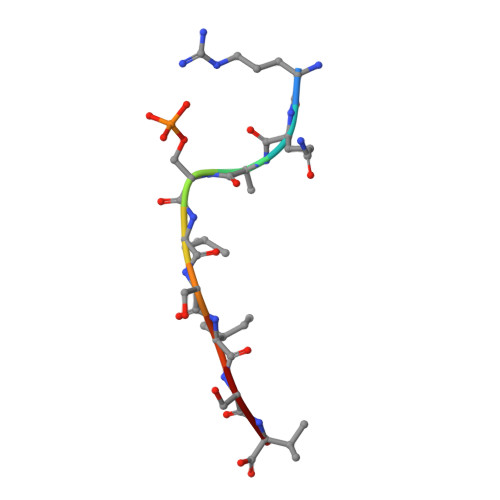
Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Mohammed, F., Cobbold, M., Zarling, A.L., Salim, M., Barrett-Wilt, G.A., Shabanowitz, J., Hunt, D.F., Engelhard, V.H., Willcox, B.E.(2008) Nat Immunol 9: 1236-1243
- PubMed: 18836451
- DOI: https://doi.org/10.1038/ni.1660
- Primary Citation of Related Structures:
3BGM, 3BH8, 3BH9, 3BHB - PubMed Abstract:
Protein phosphorylation generates a source of phosphopeptides that are presented by major histocompatibility complex class I molecules and recognized by T cells. As deregulated phosphorylation is a hallmark of malignant transformation, the differential display of phosphopeptides on cancer cells provides an immunological signature of 'transformed self'. Here we demonstrate that phosphorylation can considerably increase peptide binding affinity for HLA-A2. To understand this, we solved crystal structures of four phosphopeptide-HLA-A2 complexes. These identified a novel peptide-binding motif centered on a solvent-exposed phosphate anchor. Our findings indicate that deregulated phosphorylation can create neoantigens by promoting binding to major histocompatibility complex molecules or by affecting the antigenic identity of presented epitopes. These results highlight the potential of phosphopeptides as novel targets for cancer immunotherapy.
- Cancer Research UK Institute for Cancer Studies, School of Cancer Sciences, University of Birmingham, Edgbaston, Birmingham, UK.
Organizational Affiliation: