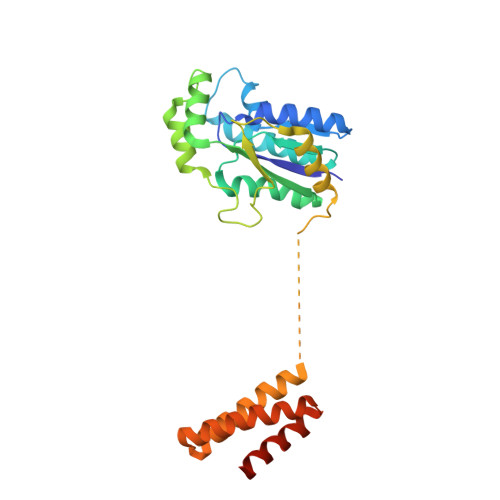
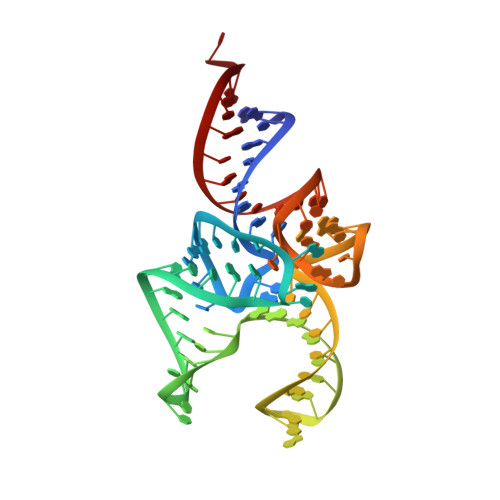
C-terminal domain of archaeal O-phosphoseryl-tRNA kinase displays large-scale motion to bind the 7-bp D-stem of archaeal tRNA(Sec)
Sherrer, R.L., Araiso, Y., Aldag, C., Ishitani, R., Ho, J.M.L., Soll, D., Nureki, O.(2011) Nucleic Acids Res 39: 1034-1041
- PubMed: 20870747
- DOI: https://doi.org/10.1093/nar/gkq845
- Primary Citation of Related Structures:
3AM1 - PubMed Abstract:
O-Phosphoseryl-tRNA kinase (PSTK) is the key enzyme in recruiting selenocysteine (Sec) to the genetic code of archaea and eukaryotes. The enzyme phosphorylates Ser-tRNA(Sec) to produce O-phosphoseryl-tRNA(Sec) (Sep-tRNA(Sec)) that is then converted to Sec-tRNA(Sec) by Sep-tRNA:Sec-tRNA synthase. Earlier we reported the structure of the Methanocaldococcus jannaschii PSTK (MjPSTK) complexed with AMPPNP. This study presents the crystal structure (at 2.4-Å resolution) of MjPSTK complexed with an anticodon-stem/loop truncated tRNA(Sec) (Mj*tRNA(Sec)), a good enzyme substrate. Mj*tRNA(Sec) is bound between the enzyme's C-terminal domain (CTD) and N-terminal kinase domain (NTD) that are connected by a flexible 11 amino acid linker. Upon Mj*tRNA(Sec) recognition the CTD undergoes a 62-Å movement to allow proper binding of the 7-bp D-stem. This large reorganization of the PSTK quaternary structure likely provides a means by which the unique tRNA(Sec) species can be accurately recognized with high affinity by the translation machinery. However, while the NTD recognizes the tRNA acceptor helix, shortened versions of MjPSTK (representing only 60% of the original size, in which the entire CTD, linker loop and an adjacent NTD helix are missing) are still active in vivo and in vitro, albeit with reduced activity compared to the full-length enzyme.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520-8114, USA.
Organizational Affiliation: