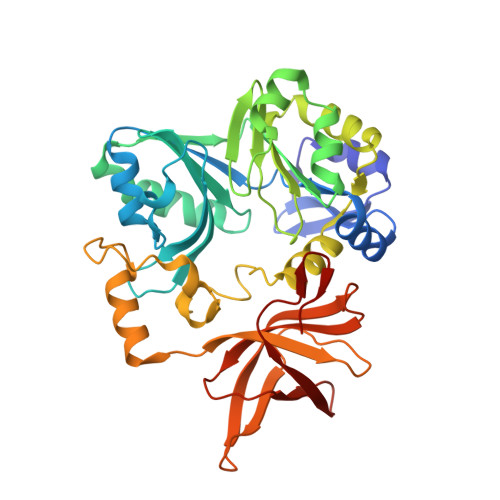
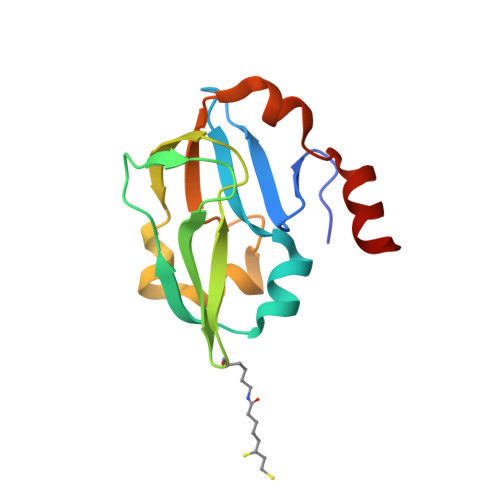
Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
Okamura-Ikeda, K., Hosaka, H., Maita, N., Fujiwara, K., Yoshizawa, A.C., Nakagawa, A., Taniguchi, H.(2010) J Biological Chem 285: 18684-18692
- PubMed: 20375021
- DOI: https://doi.org/10.1074/jbc.M110.110718
- Primary Citation of Related Structures:
3A8I, 3A8J, 3A8K, 3AB9 - PubMed Abstract:
Aminomethyltransferase, a component of the glycine cleavage system termed T-protein, reversibly catalyzes the degradation of the aminomethyl moiety of glycine attached to the lipoate cofactor of H-protein, resulting in the production of ammonia, 5,10-methylenetetrahydrofolate, and dihydrolipoate-bearing H-protein in the presence of tetrahydrofolate. Several mutations in the human T-protein gene are known to cause nonketotic hyperglycinemia. Here, we report the crystal structure of Escherichia coli T-protein in complex with dihydrolipoate-bearing H-protein and 5-methyltetrahydrofolate, a complex mimicking the ternary complex in the reverse reaction. The structure of the complex shows a highly interacting intermolecular interface limited to a small area and the protein-bound dihydrolipoyllysine arm inserted into the active site cavity of the T-protein. Invariant Arg(292) of the T-protein is essential for complex assembly. The structure also provides novel insights in understanding the disease-causing mutations, in addition to the disease-related impairment in the cofactor-enzyme interactions reported previously. Furthermore, structural and mutational analyses suggest that the reversible transfer of the methylene group between the lipoate and tetrahydrofolate should proceed through the electron relay-assisted iminium intermediate formation.
- Institute for Enzyme Research, the University of Tokushima, Tokushima 770-8503, Japan. ikeda@ier.tokushima-u.ac.jp
Organizational Affiliation: