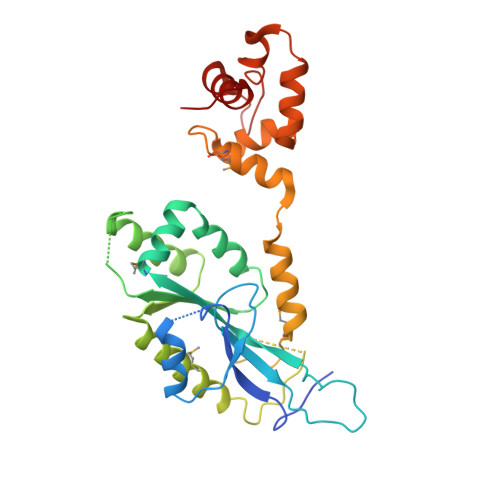
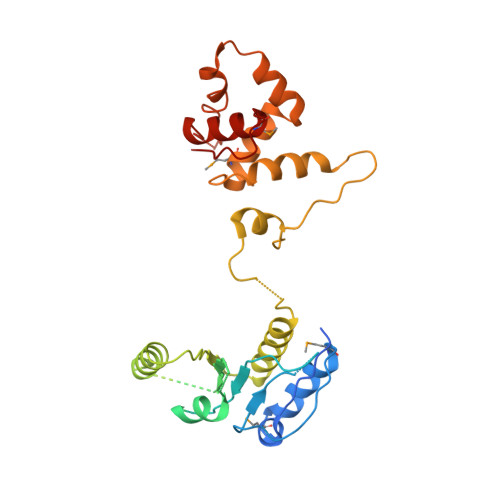
Crystal structure of the Mus81-Eme1 complex
Chang, J.H., Kim, J.J., Choi, J.M., Lee, J.H., Cho, Y.(2008) Genes Dev 22: 1093-1106
- PubMed: 18413719
- DOI: https://doi.org/10.1101/gad.1618708
- Primary Citation of Related Structures:
2ZIU, 2ZIV, 2ZIW, 2ZIX - PubMed Abstract:
The Mus81-Eme1 complex is a structure-specific endonuclease that plays an important role in rescuing stalled replication forks and resolving the meiotic recombination intermediates in eukaryotes. We have determined the crystal structure of the Mus81-Eme1 complex. Both Mus81 and Eme1 consist of a central nuclease domain, two repeats of the helix-hairpin-helix (HhH) motif at their C-terminal region, and a linker helix. While each domain structure resembles archaeal XPF homologs, the overall structure is significantly different from those due to the structure of a linker helix. We show that a flexible intradomain linker that formed with 36 residues in the nuclease domain of Eme1 is essential for the recognition of DNA. We identified several basic residues lining the outer surface of the active site cleft of Mus81 that are involved in the interaction with a flexible arm of a nicked Holliday junction (HJ). These interactions might contribute to the optimal positioning of the opposite junction across the nick into the catalytic site, which provided the basis for the "nick and counternick" mechanism of Mus81-Eme1 and for the nicked HJ to be the favored in vitro substrate of this enzyme.
- National Creative Initiatives for Structural Biology and Department of Life Science, Pohang University of Science and Technology, Pohang, KyungBook 790-784, South Korea.
Organizational Affiliation: