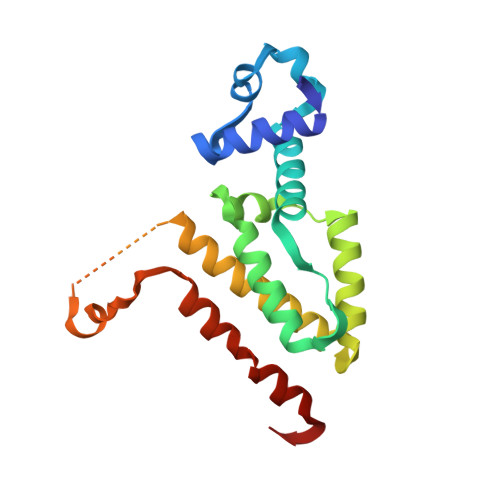
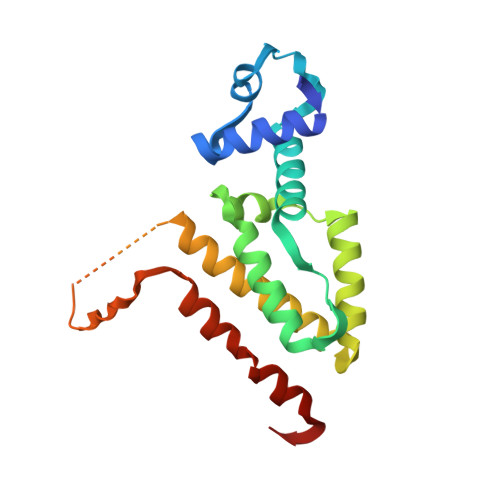
Computational Design of a Chain-Specific Tetracycline Repressor Heterodimer.
Stiebritz, M.T., Wengrzik, S., Klein, D.L., Richter, J.P., Srebrzynski, A., Weiler, S., Muller, Y.A.(2010) J Mol Biology 403: 371
- PubMed: 20816982
- DOI: https://doi.org/10.1016/j.jmb.2010.07.055
- Primary Citation of Related Structures:
2XGC, 2XGD, 2XGE - PubMed Abstract:
The specificity and selectivity of protein-protein interactions are of central importance for many biological processes, including signal transduction and transcription control. We used the in-house side-chain packing program MUMBO to computationally design a chain-specific heterodimeric variant of the bacterial transcription regulator tetracycline repressor (TetR), called T-A(A)B. Our goal was to engineer two different TetR chain variants, A and B, that no longer interact as AA or BB homodimers but selectively recombine to form heterodimers. Although 56 residues from each chain contribute to a dimer interface as large as 2200 Å(2) in wild-type TetR, the substitution of only three residues in one chain and two residues in a second chain sufficed for generating specificity in a T-A(A)B heterodimer variant. The design was corroborated in vivo by a cell-based transcription assay, and in vitro by CD spectroscopy and X-ray crystallography. Crystal structure analyses showed that while selectivity in the B chain is achieved entirely through van der Waals repulsion, the best selectivity in the A chain is obtained for the variant with the lowest number of atoms in the interface, thus possibly leading to underpacking of the dimer interface. This results in a marked decrease in thermal stability and a drastic reduction in the solubility of the T-A(A)A(A) homodimer in comparison to the designed T-A(A)B heterodimer variant.
- Lehrstuhl für Biotechnik, Department of Biology, Friedrich-Alexander University Erlangen-Nuremberg, Im IZMP, Henkestr. 91, D-91052 Erlangen, Germany.
Organizational Affiliation: