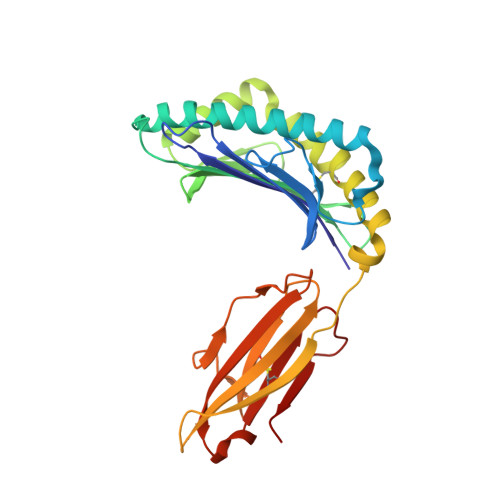
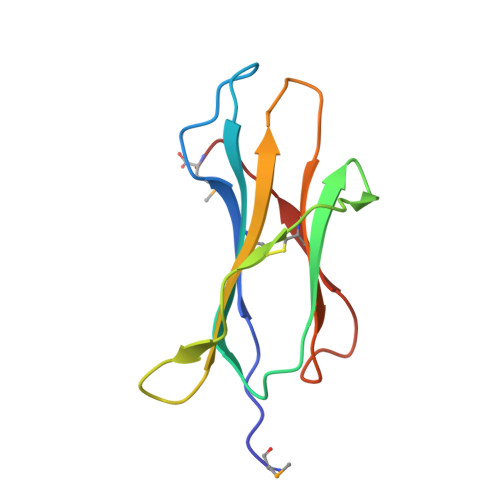
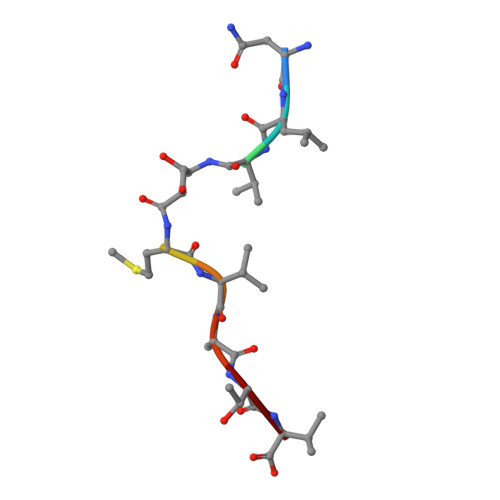
Class I Major Histocompatibility Complexes Loaded by a Periodate Trigger.
Rodenko, B., Toebes, M., Celie, P.H.N., Perrakis, A., Schumacher, T.N.M., Ovaa, H.(2009) J Am Chem Soc 131: 12305
- PubMed: 19655751
- DOI: https://doi.org/10.1021/ja9037565
- Primary Citation of Related Structures:
2X4P, 2X4Q, 2X4T - PubMed Abstract:
Class I major histocompatibility complexes (MHCs) present peptide ligands on the cell surface for recognition by appropriate cytotoxic T cells. The unstable nature of unliganded MHC necessitates the production of recombinant class I complexes through in vitro refolding reactions in the presence of an added excess of peptides. This strategy is not amenable to high-throughput production of vast collections of class I complexes. To address this issue, we recently designed photocaged MHC ligands that can be cleaved by a UV light trigger in the MHC bound state under conditions that do not affect the integrity of the MHC structure. The results obtained with photocaged MHC ligands demonstrate that conditional MHC ligands can form a generally applicable concept for the creation of defined peptide-MHCs. However, the use of UV exposure to mediate ligand exchange is unsuited for a number of applications, due to the lack of UV penetration through cell culture systems and due to the transfer of heat upon UV irradiation, which can induce evaporation. To overcome these limitations, here, we provide proof-of-concept for the generation of defined peptide-MHCs by chemical trigger-induced ligand exchange. The crystal structure of the MHC with the novel chemosensitive ligand showcases that the ligand occupies the expected binding site, in a conformation where the hydroxyl groups should be reactive to periodate. We proceed to validate this technology by producing peptide-MHCs that can be used for T cell detection. The methodology that we describe here should allow loading of MHCs with defined peptides in cell culture devices, thereby permitting antigen-specific T cell expansion and purification for cell therapy. In addition, this technology will be useful to develop miniaturized assay systems for performing high-throughput screens for natural and unnatural MHC ligands.
- Division of Cell Biology II, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Organizational Affiliation: