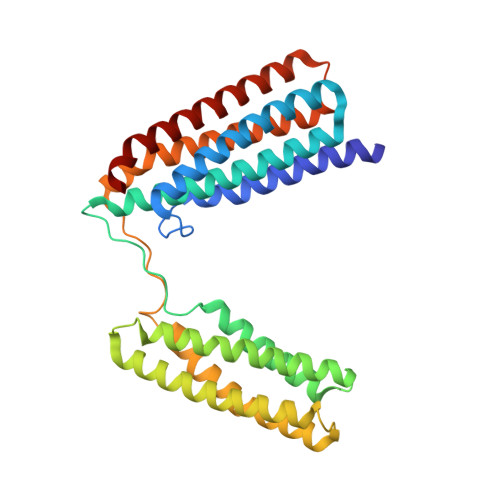
Central Region of Talin Has a Unique Fold that Binds Vinculin and Actin.
Gingras, A.R., Bate, N., Goult, B.T., Patel, B., Kopp, P.M., Emsley, J., Barsukov, I.L., Roberts, G.C.K., Critchley, D.R.(2010) J Biological Chem 285: 29577
- PubMed: 20610383
- DOI: https://doi.org/10.1074/jbc.M109.095455
- Primary Citation of Related Structures:
2X0C - PubMed Abstract:
Talin is an adaptor protein that couples integrins to F-actin. Structural studies show that the N-terminal talin head contains an atypical FERM domain, whereas the N- and C-terminal parts of the talin rod include a series of α-helical bundles. However, determining the structure of the central part of the rod has proved problematic. Residues 1359-1659 are homologous to the MESDc1 gene product, and we therefore expressed this region of talin in Escherichia coli. The crystal structure shows a unique fold comprised of a 5- and 4-helix bundle. The 5-helix bundle is composed of nonsequential helices due to insertion of the 4-helix bundle into the loop at the C terminus of helix α3. The linker connecting the bundles forms a two-stranded anti-parallel β-sheet likely limiting the relative movement of the two bundles. Because the 5-helix bundle contains the N and C termini of this module, we propose that it is linked by short loops to adjacent bundles, whereas the 4-helix bundle protrudes from the rod. This suggests the 4-helix bundle has a unique role, and its pI (7.8) is higher than other rod domains. Both helical bundles contain vinculin-binding sites but that in the isolated 5-helix bundle is cryptic, whereas that in the isolated 4-helix bundle is constitutively active. In contrast, both bundles are required for actin binding. Finally, we show that the MESDc1 protein, which is predicted to have a similar fold, is a novel actin-binding protein.
- Department of Biochemistry, University of Leicester, Lancaster Road, Leicester LE1 9HN, UK.
Organizational Affiliation: