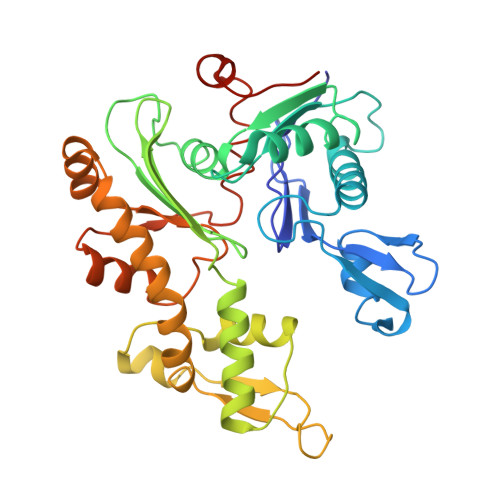
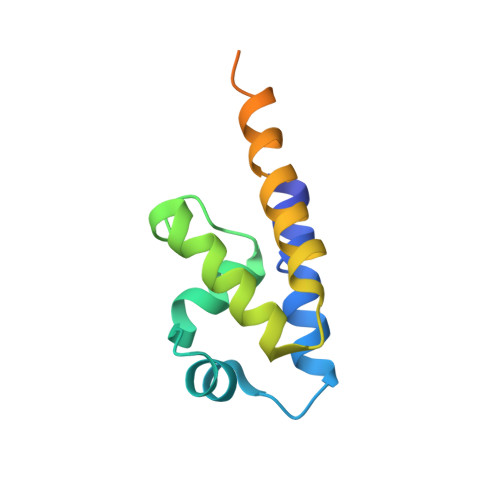
Bacterial Actin Mreb Assembles in Complex with Cell Shape Protein Rodz.
van den Ent, F., Johnson, C.M., Persons, L., deBoer, P., Lowe, J.(2010) EMBO J 29: 1081
- PubMed: 20168300
- DOI: https://doi.org/10.1038/emboj.2010.9
- Primary Citation of Related Structures:
2WUS - PubMed Abstract:
Bacterial actin homologue MreB is required for cell shape maintenance in most non-spherical bacteria, where it assembles into helical structures just underneath the cytoplasmic membrane. Proper assembly of the actin cytoskeleton requires RodZ, a conserved, bitopic membrane protein that colocalises to MreB and is essential for cell shape determination. Here, we present the first crystal structure of bacterial actin engaged with a natural partner and provide a clear functional significance of the interaction. We show that the cytoplasmic helix-turn-helix motif of Thermotoga maritima RodZ directly interacts with monomeric as well as filamentous MreB and present the crystal structure of the complex. In vitro and in vivo analyses of mutant T. maritima and Escherichia coli RodZ validate the structure and reveal the importance of the MreB-RodZ interaction in the ability of cells to propagate as rods. Furthermore, the results elucidate how the bacterial actin cytoskeleton might be anchored to the membrane to help constrain peptidoglycan synthesis in the periplasm.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: