Identification of Novel Allosteric Inhibitors of Hepatitis C Virus Ns5B Polymerase Thumb Domain (Site II) by Structure-Based Design
Di Francesco, M.E., Di Marco, S., Summa, V.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA-DIRECTED RNA POLYMERASE | 536 | Hepacivirus hominis | Mutation(s): 0 EC: 2.7.7.48 | 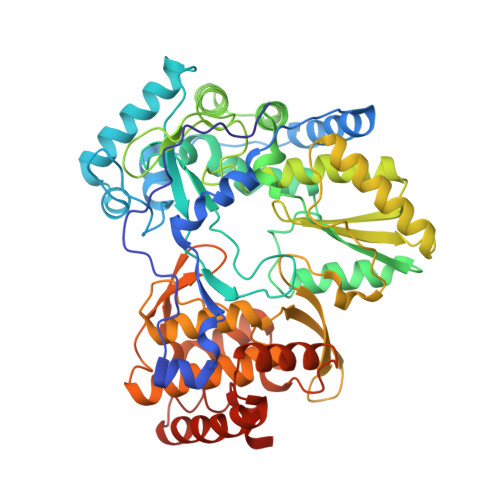 | |
UniProt | |||||
Find proteins for P26663 (Hepatitis C virus genotype 1b (isolate BK)) Explore P26663 Go to UniProtKB: P26663 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P26663 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| QQ3 Query on QQ3 | B [auth A], C [auth A], D [auth A] | (3R)-3-(4-METHYL-1,3-DIOXO-1,3-DIHYDRO-2H-PYRROLO[3,4-C]QUINOLIN-2-YL)HEXANOIC ACID C18 H18 N2 O4 AZYBNXOGXCANBA-LLVKDONJSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 66.764 | α = 90 |
| b = 96.175 | β = 90 |
| c = 95.683 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| AMoRE | phasing |