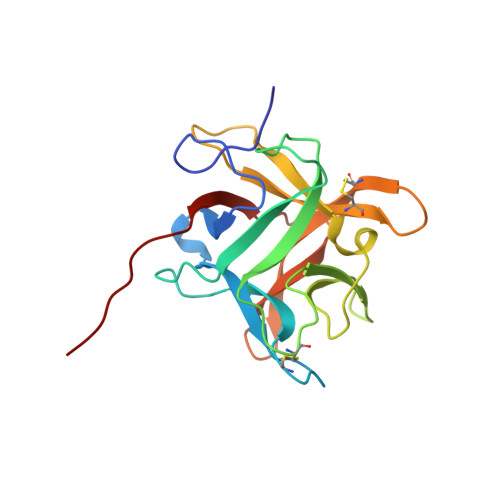
Refined crystal structure (2.3 A) of a double-headed winged bean alpha-chymotrypsin inhibitor and location of its second reactive site.
Dattagupta, J.K., Podder, A., Chakrabarti, C., Sen, U., Mukhopadhyay, D., Dutta, S.K., Singh, M.(1999) Proteins 35: 321-331
- PubMed: 10328267
- DOI: https://doi.org/10.1002/(sici)1097-0134(19990515)35:3<321::aid-prot6>3.0.co;2-y
- Primary Citation of Related Structures:
2WBC - PubMed Abstract:
The crystal structure of a double-headed alpha-chymotrypsin inhibitor, WCI, from winged bean seeds has now been refined at 2.3 A resolution to an R-factor of 18.7% for 9,897 reflections. The crystals belong to the hexagonal space group P6(1)22 with cell parameters a = b = 61.8 A and c = 212.8 A. The final model has a good stereochemistry and a root mean square deviation of 0.011 A and 1.14 degrees from ideality for bond length and bond angles, respectively. A total of 109 ordered solvent molecules were localized in the structure. This improved structure at 2.3 A led to an understanding of the mechanism of inhibition of the protein against alpha-chymotrypsin. An analysis of this higher resolution structure also helped us to predict the location of the second reactive site of the protein, about which no previous biochemical information was available. The inhibitor structure is spherical and has twelve anti-parallel beta-strands with connecting loops arranged in a characteristic beta-trefoil fold common to other homologous serine protease inhibitors in the Kunitz (STI) family as well as to some non homologous functionally unrelated proteins. A wide variation in the surface loop regions is seen in the latter ones.
- Crystallography and Molecular Biology Division, Saha Institute of Nuclear Physics, Calcutta, India. jiban@cmb2.saha.ernet.in
Organizational Affiliation: